The nucleoporin Nup188 controls passage of membrane proteins across the nuclear pore complex - PubMed (original) (raw)
The nucleoporin Nup188 controls passage of membrane proteins across the nuclear pore complex
Gandhi Theerthagiri et al. J Cell Biol. 2010.
Abstract
All transport across the nuclear envelope (NE) is mediated by nuclear pore complexes (NPCs). Despite their enormous size, approximately 60 MD in vertebrates, they are comprised of only approximately 30 distinct proteins (nucleoporins or Nups), many of which form subcomplexes that act as building blocks for NPC assembly. One of these evolutionarily conserved subcomplexes, the Nup93 complex, is a major structural component linking the NPC to the membranes of the NE. Using in vitro nuclear assembly assays, we show that two components of the Nup93 complex, Nup188 and Nup205, are dispensable for NPC formation. However, nuclei lacking Nup188 increase in size by several fold compared with wild type. We demonstrate that this phenotype is caused by an accelerated translocation of integral membrane proteins through NPCs, suggesting that Nup188 confines the passage of membrane proteins and is thus crucial for the homeostasis of the different nuclear membranes.
Figures
Figure 1.
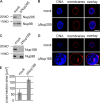
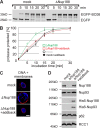
Nup93 forms two distinct complexes. (A) Immunoprecipitations were performed from Xenopus egg cytosol with antibodies against Nup188 (two antibodies raised in different rabbits), Nup93, Nup205, Nup155, and Nup53 and, as a control, Nup98. The indicated proteins were identified by Western blotting with the respective antisera. 10% of starting material (input) was loaded on the left. (B) Quantitative immunoprecipitations were performed from limited amounts of Xenopus egg cytosol with antibodies against Nup188, Nup93, and Nup205 and, as a control, Nup98. Immunoprecipitates and unbound material as well as 10, 30, and 100% of the starting material (input) were analyzed by Western blotting. The Nup93 antibody also recognized a cross-reactivity (asterisk) in the starting and unbound material migrating slightly slower.
Figure 2.
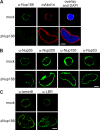
Removal of Nup188–Nup93 enlarges nuclei. (A and C) Western blot analysis of mock- and Nup205–Nup93 (A)- or Nup188–Nup93 (C)-depleted extracts. (B and D) Nuclei were assembled in mock- and Nup205–Nup93 (B)- or Nup188–Nup93 (D)-depleted extracts for 90 min, fixed with 4% PFA and 0.5% glutaraldehyde, and analyzed for chromatin and membrane staining (blue, DAPI; red, DiIC18). (E) Quantitation of the cross-sectional area of mock- and Nup188–Nup93-depleted nuclei of experiments performed as in D. More than 100 randomly chosen chromatin substrates were counted per reaction. The mean of three independent experiments is shown, and error bars represent the total variation. Bars, 20 µm.
Figure 3.
Nup188–Nup93-depleted nuclei contain NPCs and INM proteins. (A) Nuclei were assembled in mock- or Nup188–Nup93-depleted extracts for 90 min, fixed with 4% PFA, and analyzed with Nup188-specific antibodies (green) or the monoclonal antibody mAb414, which recognizes FG-repeat nucleoporins (red). Chromatin is stained with DAPI. (B and C) Samples were prepared as in A and analyzed by immunofluorescence with the indicated antibodies. Bars, 20 µm.
Figure 4.
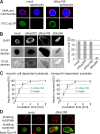
Nuclear functions are unaffected by Nup188–Nup93 depletion. (A) Nuclei were assembled in mock- or Nup188–Nup93-depleted extracts. After 50 min, fluorescently labeled dUTP (green in overlay) and, where indicated, aphidicolin were added. After 90 min, replication was analyzed by confocal microscopy. Membranes were stained with DiIC18 (red), and chromatin was stained with DAPI (blue). (B) Nuclei were assembled in mock-, Nup205–Nup93-, Nup188–Nup93-, and Nup98-depleted extracts. After 90 min, fluorescently labeled 2-MD (not depicted), 70-kD (middle), and 10-kD (bottom) dextrans were added. DNA was stained with DAPI, and samples were analyzed by confocal microscopy. For quantitation, only nuclei with an intact NE (as judged by the exclusion of 2-MD dextrans; >95% of nuclei in each sample) were analyzed. For mock- and Nup188–Nup93-depleted samples, seven independent experiments were analyzed (error bars show the standard deviation), and for Nup205–Nup93- and Nup98-depleted samples, two independent experiments were analyzed. (C) Nuclei were assembled as in A. After 50 min, i.e., when a closed NE had formed, importin α/β– or transportin-dependent reporter substrates containing a TEV cleavage site were added. To assay the nuclear import kinetics of the reporter, NusA-fused TEV protease was added at the indicated time points. Protease cleavage was stopped after 1 min by the addition of SDS sample buffer and boiling. Lines mark the mean of four independent experiments. (D) Nuclei were assembled as in A, and an EGFP-fused shuttling substrate was added. Nuclear export function was inhibited by the addition of 300 nM leptomycin B. Nuclei were isolated and analyzed by confocal microscopy. Membranes are stained with DiIC18 (red). Bars, 20 µm.
Figure 5.
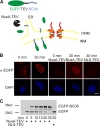
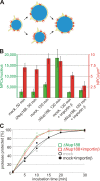
An assay for the transport of membrane proteins to the INM. (A) Schematic representation of the assay. The transmembrane-containing INM protein BC08 is fused to an EGFP domain followed by a recognition site for the TEV protease (top). The fusion protein is reconstituted into liposomes that rapidly fuse with the ER and ONM in the nuclear assembly reaction. INM localization of the reporter can be distinguished from ONM and ER localization by the reporter’s protection from TEV protease fused with a large NusA domain to prevent its diffusion through the NPC. (B) Nuclei were assembled in Xenopus egg extracts. After 50 min, i.e., when a closed NE had formed, proteoliposomes containing the reporter were added. At the indicated time points, buffer, NusA-fused TEV protease, or TEV protease linked to NLS peptides was added. Protease cleavage was stopped after 5 min by fixation, and samples were processed for immunofluorescence and analyzed by confocal microscopy. α-EGFP immunofluorescence is shown in red, and DAPI is shown in blue. (C) Reactions were performed as in B. Protease cleavage was stopped by the addition of SDS sample buffer and boiling. Protease protection of the reporter was analyzed by Western blotting with the position of the uncleaved (EGFP-BC08) and the cleaved reporter with only the EGFP visible indicated. Bar, 20 µm.
Figure 6.
Nup188–Nup93 depletion causes a faster delivery of integral membrane proteins to the INM. (A) Nuclei were assembled in mock- or Nup188–Nup93-depleted Xenopus egg extracts and analyzed by Western blotting. Positions of the uncleaved (EGFP-BC08) and the cleaved (EGFP) reporter are indicated. (B) Quantification from three independent experiments performed as in A. Squares are from Nup188–Nup93-depleted samples (green without and red with the addition of in vitro translated Nup188–Nup93 [addback]), and circles are from mock-treated samples. Lines mark the mean of three independent experiments. (C) The addition of recombinant Nup188–Nup93 rescues the Nup188–Nup93 depletion phenotype. Nuclei were assembled for 120 min in mock- or Nup188–Nup93-depleted extracts without or with the addition of in vitro translated Nup188–Nup93 (addback), fixed with 4% PFA and 0.5% glutaraldehyde, and analyzed for chromatin and membrane staining (blue, DAPI; red, DiIC18). (D) In vitro translated Nup188–Nup93 is incorporated into in vitro assembled nuclei. Nuclei were assembled for 1 h to ensure approximately equal NPC numbers per nucleus, isolated, and analyzed by Western blotting. In vitro translated Nup188 and Nup93 could be also detected using an anti-His6 antibody (third and fourth row). The nucleoporin p62 was detected with mAb414, and the chromatin-binding protein RCC1 serves as a control for equal loading of nuclei. Bar, 20 µm.
Figure 7.
The nuclear growth phenotype and the faster INM targeting are independent of the increased NPC number upon Nup188–Nup93 depletion. (A) Two possible scenarios for the relationship of the nuclear membrane expansion and the increase in NPC numbers upon Nup188–Nup93 depletion. In the top route, Nup188–Nup93 depletion causes a faster growth of the NE, which in turn allows more NPCs (red) to be integrated. In the bottom route, NPC assembly increases on Nup188–Nup93-depleted nuclei. The resulting higher total transport capacity of these allows accelerated nuclear volume and envelope expansion. (B) Nuclei were assembled in mock- and Nup188–Nup93-depleted extracts for 50 min and 120 min. Where indicated, de novo NPC assembly was blocked by the addition of 2 µM importin β after 50 min, i.e., after a closed NE had formed, and nuclei were further incubated for 70 min. NPC numbers per nucleus (green bars) and NPC density (red bars) were quantified. (C) Nuclei were assembled in mock- and Nup188–Nup93-depleted extracts for 50 min. Then, where indicated, de novo NPC assembly was blocked by the addition of 2 µM importin β. INM targeting of the reporter was analyzed as in Fig. 6. Squares are from Nup188–Nup93-depleted samples, and circles from mock-treated samples. Lines mark the mean of two independent experiments.
Similar articles
- The C-terminal domain of Nup93 is essential for assembly of the structural backbone of nuclear pore complexes.
Sachdev R, Sieverding C, Flötenmeyer M, Antonin W. Sachdev R, et al. Mol Biol Cell. 2012 Feb;23(4):740-9. doi: 10.1091/mbc.E11-09-0761. Epub 2011 Dec 14. Mol Biol Cell. 2012. PMID: 22171326 Free PMC article. - NEP-A and NEP-B both contribute to nuclear pore formation in Xenopus eggs and oocytes.
Salpingidou G, Rzepecki R, Kiseleva E, Lyon C, Lane B, Fusiek K, Golebiewska A, Drummond S, Allen T, Ellis JA, Smythe C, Goldberg MW, Hutchison CJ. Salpingidou G, et al. J Cell Sci. 2008 Mar 1;121(Pt 5):706-16. doi: 10.1242/jcs.019968. Epub 2008 Feb 12. J Cell Sci. 2008. PMID: 18270266 - Caenorhabditis elegans nucleoporins Nup93 and Nup205 determine the limit of nuclear pore complex size exclusion in vivo.
Galy V, Mattaj IW, Askjaer P. Galy V, et al. Mol Biol Cell. 2003 Dec;14(12):5104-15. doi: 10.1091/mbc.e03-04-0237. Epub 2003 Aug 22. Mol Biol Cell. 2003. PMID: 12937276 Free PMC article. - Structure and Assembly of the Nuclear Pore Complex.
Hampoelz B, Andres-Pons A, Kastritis P, Beck M. Hampoelz B, et al. Annu Rev Biophys. 2019 May 6;48:515-536. doi: 10.1146/annurev-biophys-052118-115308. Epub 2019 Apr 3. Annu Rev Biophys. 2019. PMID: 30943044 Review. - Functional architecture of the nuclear pore complex.
Grossman E, Medalia O, Zwerger M. Grossman E, et al. Annu Rev Biophys. 2012;41:557-84. doi: 10.1146/annurev-biophys-050511-102328. Annu Rev Biophys. 2012. PMID: 22577827 Review.
Cited by
- Building a nuclear envelope at the end of mitosis: coordinating membrane reorganization, nuclear pore complex assembly, and chromatin de-condensation.
Schooley A, Vollmer B, Antonin W. Schooley A, et al. Chromosoma. 2012 Dec;121(6):539-54. doi: 10.1007/s00412-012-0388-3. Epub 2012 Oct 27. Chromosoma. 2012. PMID: 23104094 Free PMC article. Review. - FG repeats facilitate integral protein trafficking to the inner nuclear membrane.
Kerr AR, Schirmer EC. Kerr AR, et al. Commun Integr Biol. 2011 Sep;4(5):557-9. doi: 10.4161/cib.4.5.16052. Epub 2011 Sep 1. Commun Integr Biol. 2011. PMID: 22046461 Free PMC article. - Sizing and shaping the nucleus: mechanisms and significance.
Jevtić P, Edens LJ, Vuković LD, Levy DL. Jevtić P, et al. Curr Opin Cell Biol. 2014 Jun;28:16-27. doi: 10.1016/j.ceb.2014.01.003. Epub 2014 Feb 4. Curr Opin Cell Biol. 2014. PMID: 24503411 Free PMC article. Review. - Differential turnover of Nup188 controls its levels at centrosomes and role in centriole duplication.
Vishnoi N, Dhanasekeran K, Chalfant M, Surovstev I, Khokha MK, Lusk CP. Vishnoi N, et al. J Cell Biol. 2020 Mar 2;219(3):e201906031. doi: 10.1083/jcb.201906031. J Cell Biol. 2020. PMID: 32211895 Free PMC article. - Nuclear size is regulated by importin α and Ntf2 in Xenopus.
Levy DL, Heald R. Levy DL, et al. Cell. 2010 Oct 15;143(2):288-98. doi: 10.1016/j.cell.2010.09.012. Cell. 2010. PMID: 20946986 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources