Hidden Markov model speed heuristic and iterative HMM search procedure - PubMed (original) (raw)
Hidden Markov model speed heuristic and iterative HMM search procedure
L Steven Johnson et al. BMC Bioinformatics. 2010.
Abstract
Background: Profile hidden Markov models (profile-HMMs) are sensitive tools for remote protein homology detection, but the main scoring algorithms, Viterbi or Forward, require considerable time to search large sequence databases.
Results: We have designed a series of database filtering steps, HMMERHEAD, that are applied prior to the scoring algorithms, as implemented in the HMMER package, in an effort to reduce search time. Using this heuristic, we obtain a 20-fold decrease in Forward and a 6-fold decrease in Viterbi search time with a minimal loss in sensitivity relative to the unfiltered approaches. We then implemented an iterative profile-HMM search method, JackHMMER, which employs the HMMERHEAD heuristic. Due to our search heuristic, we eliminated the subdatabase creation that is common in current iterative profile-HMM approaches. On our benchmark, JackHMMER detects 14% more remote protein homologs than SAM's iterative method T2K.
Conclusions: Our search heuristic, HMMERHEAD, significantly reduces the time needed to score a profile-HMM against large sequence databases. This search heuristic allowed us to implement an iterative profile-HMM search method, JackHMMER, which detects significantly more remote protein homologs than SAM's T2K and NCBI's PSI-BLAST.
Figures
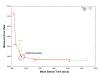
Figure 1
Minimum Error Rate Relative to Mean Search Time. 250 randomly chosen benchmarking models were used in HMMERHEAD Viterbi searches of NRDB90 and the test database utilizing a range of θ and η values. All other parameters were kept at their default value (θ = 6, δ = 2, μ = 7, and η = 20). Plotting minimum error rate versus mean search time for searches using these parameter values reveals a dramatic increase in minimum error rate, relative to a minor decrease in search time, for θ and η parameter values greater than 6 and 20, respectively. This further supports our choice of these parameter values for HMMERHEAD's default settings. The total number of true homologous pairs between those 250 models and the test benchmark was 3,617. The number of true positives identified at these parameter settings at 0 false positives is 938 or 26% of the possible true positives.
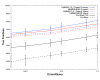
Figure 2
Remote Homology Detection of HMMERHEAD and HMMER 2.5.1 Forward and Viterbi. Each of the 2,521 benchmarking models was scored against the test database using either HMMER 2.5.1 or HMMERHEAD with Forward or Viterbi scoring. The results of the searches were combined and scored. This procedure was then repeated for each of the 1000 bootstrapping replicate test sets. The average number of true positives was plotted versus errors per query. Minimum and maximum numbers of true positives from the replicates are shown as error bars. HMMERHEAD Forward (Red) performance is shown relative to default HMMER 2.5.1 Forward (Blue), WU-BLAST (Black dashed), and WU-BLAST Family Pairwise Search (Black). Default HMMERHEAD Forward detects an average of 269 fewer true positives than default HMMER 2.5.1 Forward and detects significantly more true positives than either pairwise sequence comparison method. HMMERHEAD Viterbi (Red dashed) performance is shown relative to default HMMER 2.5.1 Viterbi (Blue dashed). Default HMMERHEAD Viterbi detects an average of 173 fewer true positives than default HMMER 2.5.1 Viterbi and again outperforms either pairwise comparison method. The total number of true homologous pairs between the 2,521 models and the test database is 39,733, and thus 8,000 true positives correspond to identifying 20% of the homologs.
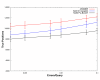
Figure 3
Performance of Iterative Methods. The individual 2,521 benchmarking sequences were used to iteratively search a non-redundant version of NCBI's NR database. The iterative models created from this process were then scored against the test database. The results of the searches were combined and scored. This procedure was then repeated for each of the 1000 bootstrapping replicate testsets. The average number of true positives was plotted versus errors per query. Minimum and maximums numbers of true positives from the replicates are shown as error bars. JackHMMER (Red) detects an average of 1,337 more homologs than SAM 3.5's target2k (Blue) and an average of 2,476 more homologs than NCBI's PSI-BLAST (Black) across the errors per query range. This represents an increase of 14% and 28% in remote protein homologs detected. As before, the total number of true homologous pairs between the 2,521 models and the test database is 39,733, and thus 12,000 true positives correspond to identifying 30% of the homologs.
Similar articles
- Accelerated Profile HMM Searches.
Eddy SR. Eddy SR. PLoS Comput Biol. 2011 Oct;7(10):e1002195. doi: 10.1371/journal.pcbi.1002195. Epub 2011 Oct 20. PLoS Comput Biol. 2011. PMID: 22039361 Free PMC article. - Protein homology detection by HMM-HMM comparison.
Söding J. Söding J. Bioinformatics. 2005 Apr 1;21(7):951-60. doi: 10.1093/bioinformatics/bti125. Epub 2004 Nov 5. Bioinformatics. 2005. PMID: 15531603 - A comparison of profile hidden Markov model procedures for remote homology detection.
Madera M, Gough J. Madera M, et al. Nucleic Acids Res. 2002 Oct 1;30(19):4321-8. doi: 10.1093/nar/gkf544. Nucleic Acids Res. 2002. PMID: 12364612 Free PMC article. - Profile hidden Markov models.
Eddy SR. Eddy SR. Bioinformatics. 1998;14(9):755-63. doi: 10.1093/bioinformatics/14.9.755. Bioinformatics. 1998. PMID: 9918945 Review. - Utilizing profile hidden Markov model databases for discovering viruses from metagenomic data: a comprehensive review.
Yu R, Huang Z, Lam TYC, Sun Y. Yu R, et al. Brief Bioinform. 2024 May 23;25(4):bbae292. doi: 10.1093/bib/bbae292. Brief Bioinform. 2024. PMID: 39003531 Free PMC article. Review.
Cited by
- Structure prediction and analysis of DNA transposon and LINE retrotransposon proteins.
Abrusán G, Zhang Y, Szilágyi A. Abrusán G, et al. J Biol Chem. 2013 May 31;288(22):16127-38. doi: 10.1074/jbc.M113.451500. Epub 2013 Mar 25. J Biol Chem. 2013. PMID: 23530042 Free PMC article. - Enhancing alphafold-multimer-based protein complex structure prediction with MULTICOM in CASP15.
Liu J, Guo Z, Wu T, Roy RS, Quadir F, Chen C, Cheng J. Liu J, et al. Commun Biol. 2023 Nov 10;6(1):1140. doi: 10.1038/s42003-023-05525-3. Commun Biol. 2023. PMID: 37949999 Free PMC article. - Comparative Transcriptome Analysis of Two Contrasting Soybean Varieties in Response to Aluminum Toxicity.
Zhao L, Cui J, Cai Y, Yang S, Liu J, Wang W, Gai J, Hu Z, Li Y. Zhao L, et al. Int J Mol Sci. 2020 Jun 17;21(12):4316. doi: 10.3390/ijms21124316. Int J Mol Sci. 2020. PMID: 32560405 Free PMC article. - The Phytophthora sojae effector PsFYVE1 modulates immunity-related gene expression by targeting host RZ-1A protein.
Lu X, Yang Z, Song W, Miao J, Zhao H, Ji P, Li T, Si J, Yin Z, Jing M, Shen D, Dou D. Lu X, et al. Plant Physiol. 2023 Feb 12;191(2):925-945. doi: 10.1093/plphys/kiac552. Plant Physiol. 2023. PMID: 36461945 Free PMC article. - Proteome variation among Filifactor alocis strains.
Aruni AW, Roy F, Sandberg L, Fletcher HM. Aruni AW, et al. Proteomics. 2012 Nov;12(22):3343-64. doi: 10.1002/pmic.201200211. Proteomics. 2012. PMID: 23008013 Free PMC article.
References
- HMMER: biosequence analysis using profile hidden Markov models. http://hmmer.org/
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;3:403–410. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials