TDP-43-mediated neuron loss in vivo requires RNA-binding activity - PubMed (original) (raw)
TDP-43-mediated neuron loss in vivo requires RNA-binding activity
Aaron Voigt et al. PLoS One. 2010.
Abstract
Alteration and/or mutations of the ribonucleoprotein TDP-43 have been firmly linked to human neurodegenerative diseases, including amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD). The relative impacts of TDP-43 alteration, mutation, or inherent protein function on neural integrity, however, remain less clear--a situation confounded by conflicting reports based on transient and/or random-insertion transgenic expression. We therefore performed a stringent comparative investigation of impacts of these TDP-43 modifications on neural integrity in vivo. To achieve this, we systematically screened ALS/FTLD-associated and synthetic TDP-43 isoforms via same-site gene insertion and neural expression in Drosophila; followed by transposon-based motor neuron-specific transgenesis in a chick vertebrate system. Using this bi-systemic approach we uncovered a requirement of inherent TDP-43 RNA-binding function--but not ALS/FTLD-linked mutation, mislocalization, or truncation--for TDP-43-mediated neurotoxicity in vivo.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
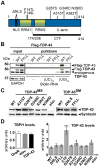
Figure 1. Analysis of different TDP-43 variants used for ectopic expression.
(A) Schematic overview of TDP-43 variants tested in vivo. Positions and nature of amino acid substitutions and size of two C-terminal fragments (CTFs) analyzed are indicated. Synthetic mutations (SM) were introduced to interfere with TDP-43 localization or function. Mutations in the nuclear localization signal (ΔNLS) were introduced to target TDP-43 to the cytoplasm and mutations F147L/F149L (FFLL) in the first RNA recognition motive (RRM1) were introduced to impair TDP-43 inherent RNA-binding capacity. (B) Loss of RNA-binding by TDP-43FFLL. Biotinylated oligonucleotides were incubated with lysates from either untransfected HEK293 cells or HEK293 cells transfected with FLAG-tagged TDP-43WT or TDP-43FFLL (left, input) followed by UV crosslinking and streptavidin-pulldown of Biotin-RNA (right, pulldown). Precipitates were separated, blotted and membranes probed with FLAG- (upper panel) and TDP-43-specific (lower panel) antibodies to assay co-precipitation of TDP-43. Note: Co-precipitation of endogenous TDP-43 with (UG)12-repeats in all lysates. Ectopic FLAG-TDP-43WT, but not FLAG-TDP-43FFLL co-precipitated with cognate UG repeats. GAPDH served as loading control for protein input. (C) Equalized pan-neural expression of _φ-C31_-inserted TDP-43 variants in adult Drosophilae. TDP-43 expression was visualized using an anti-human TDP-43 antibody. Syntaxin served as loading control. (D) Assessment of relative TBPH and TDP-43 expression levels. Left graph: Relative abundance of endogenous TBPH transcripts in relation to actin5C of wild type strain OregonR (OreR), the Gal4-driver line (elavC155::Gal4) and flies with pan-neural TDP-43 expression (elavC155::Gal4/Y;UAS::TDP-43WT/+). TBPH levels were not significantly different in analyzed genotypes. Right graph: mRNA abundance of TDP-43 in flies with pan-neural expression of the different TDP-43 variants normalized to actin5C and TBPH mRNA levels. Significant differences are indicated. *p<0.05; ***p<0.001. Head lysates of flies with pan-neural expression of TDP-43 were used for Western blot and qPCR analysis. elav::Gal4 flies without a TDP-43 transgene (elav) were used as negative control.
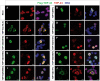
Figure 2. Localization of TDP-43 variants in human cells.
HEK293E cells transfected with N-terminal Flag-tagged TDP-43 variants detected with Flag- (left, green) and TDP-43 (middle, red) specific antibodies. Overlay (right) with Hoechst stained DNA (blue). (A) Exogenous TDP-43 had to be visualized via fused Flag-tag, as HEK cells show robust endogenous expression of TDP-43. (B) Ectopic TDP-43WT mainly localized to the nucleus. (C) In contrast, TDP-43ΔNLS exclusively localized to cytosol without showing nuclear Flag-signal. Similar to TDP-43FFLL (D), also TDP-43MS (E–I) displayed a nuclear localization. However, TDP-43FFLL localized in a characteristic punctuate pattern throughout the nucleus (D). (J) TDP-43CTF displayed a predominantly cytoplasmic localization similar to TDP-43ΔNLS (compare C and J). Scale bar indicates 10 µm.
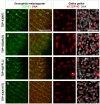
Figure 3. Localization of TDP-43 in Drosophila melanogaster and Gallus gallus.
Confocal sections of eye imaginal discs from Drosophila larvae (left panel) and motor neurons from Gallus (right panel) expressing indicated TDP-43 variants. To be able to discriminate between the two in vivo systems, ectopic TDP-43 in Drosophila is shown in green, whereas TDP-43 in Gallus is shown in red. Subcellular localization of the different TDP-43 variants was found to be identical between fly and chick. TDP-43WT (A) localized mainly to the nucleus, while TDP-43ΔNLS (B) was found predominantly in the cytoplasm. TDP-43FFLL (C) and TDP-43A315T (D) displayed a nuclear distribution. Only cells with very high expression levels of usually nuclear TDP-43 displayed a detectable cytoplasmic staining (example in case of TDP-43FFLL marked by asterisk). DNA was stained with Sytox® Orange (fly, red) or DAPI (chick, white). Scale bar indicates 50 µm. Neuronal expression was mediated by elav::Gal4 (flies) or Hb9::Cre (chick).
Figure 4. Requirement for RNA-binding activity for TDP-43-mediated neural defects in Drosophila.
(A) TDP-43 expression reduces longevity in Drosophila. Flies with pan-neural (elav::Gal4) expression of indicated TDP-43 variants were assayed for longevity. (B) Median survival of respective survival curves. (C) Cross-comparison of survival curves with regard to statistical significance after Bonferroni correction. (D) Age-dependent locomotion defects after TDP-43 expression in motor neurons. Flies expressing indicated TDP-43 variants specifically in motor neurons (D42::Gal4) were assayed for negative geotaxis at 1, 10 and 20 days post eclosion. (E) Detailed summary of statistical analysis (2-Way ANOVA followed by Bonferroni post-hoc tests) of age-dependent locomotion effects. Genotypes of flies analyzed: elavC155::Gal4/Y;UAS::TDP-43/+ in longevity analysis and w/Y;UAS::TDP-43/+;D42::Gal4/+ in case of locomotion assay. elavC155::Gal4/Y (longevity) and w/Y;;D42::Gal4 (climbing) served as controls. *p<0.05; **p<0.01; ***p<0.001; ns not significant.
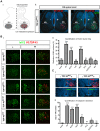
Figure 5. RNA-binding activity is required for TDP-43-mediated motor neuron loss in chick.
(A) Stable unilateral expression of human TDP-43 variants in chick (Gallus gallus) spinal cord. [i] Schematic of expression system mediating motor neuron-restricted expression upon unilateral transfection. [ii] Unilateral expression of human TDP-43FFLL (red) in embryonic day 9 (E9) chick spinal cord (nuclei labeled with DAPI: blue): transversal section at lumbar levels. “−” and “+” respectively indicate control and transfected hemicords. Isl1/2 labels motor neuron nuclei (green). (B) [i] Examples of thoracic motor columns (Isl1/2+ motor neurons: green) upon TDP-43 variant expression (red). [ii] Quantification of motor neuron loss upon TDP-43 variant expression over all obtained sections (in “-” versus “+” hemicord). Differences relative to control (t-student's test) are indicated. (C) [i] Activated Caspase-3 (green) detected in E5 motor neurons upon TDP-43WT expression (indicated by IRES-cherry bi-cistronic reporter: red). Compared to E5 motor neurons expressing TDP-43WT, little activation of Caspase-3 was detected upon TDP-43CTF expression. [ii] Quantification of Caspase-3 activation in motor neurons of transfected hemicords versus vector control. Significant differences are indicated (t-student's test - relative to control). *p<0.05; **p<0.01; ***p<0.001; ns not significant.
Similar articles
- Neuronal function and dysfunction of Drosophila dTDP.
Lin MJ, Cheng CW, Shen CK. Lin MJ, et al. PLoS One. 2011;6(6):e20371. doi: 10.1371/journal.pone.0020371. Epub 2011 Jun 1. PLoS One. 2011. PMID: 21673800 Free PMC article. - TDP-43 mutations causing amyotrophic lateral sclerosis are associated with altered expression of RNA-binding protein hnRNP K and affect the Nrf2 antioxidant pathway.
Moujalled D, Grubman A, Acevedo K, Yang S, Ke YD, Moujalled DM, Duncan C, Caragounis A, Perera ND, Turner BJ, Prudencio M, Petrucelli L, Blair I, Ittner LM, Crouch PJ, Liddell JR, White AR. Moujalled D, et al. Hum Mol Genet. 2017 May 1;26(9):1732-1746. doi: 10.1093/hmg/ddx093. Hum Mol Genet. 2017. PMID: 28334913 - Lower motor neuron involvement in TAR DNA-binding protein of 43 kDa-related frontotemporal lobar degeneration and amyotrophic lateral sclerosis.
Riku Y, Watanabe H, Yoshida M, Tatsumi S, Mimuro M, Iwasaki Y, Katsuno M, Iguchi Y, Masuda M, Senda J, Ishigaki S, Udagawa T, Sobue G. Riku Y, et al. JAMA Neurol. 2014 Feb;71(2):172-9. doi: 10.1001/jamaneurol.2013.5489. JAMA Neurol. 2014. PMID: 24378564 - Mechanisms Associated with TDP-43 Neurotoxicity in ALS/FTLD.
Shenouda M, Zhang AB, Weichert A, Robertson J. Shenouda M, et al. Adv Neurobiol. 2018;20:239-263. doi: 10.1007/978-3-319-89689-2_9. Adv Neurobiol. 2018. PMID: 29916022 Review. - [Clinical and pathological spectrum of TDP-43 associated ALS].
Onodera O, Yokoseki A, Tan CF, Ishihara T, Nishiira Y, Toyoshima Y, Kakita A, Nishizawa M, Takahashi H. Onodera O, et al. Rinsho Shinkeigaku. 2010 Nov;50(11):940-2. doi: 10.5692/clinicalneurol.50.940. Rinsho Shinkeigaku. 2010. PMID: 21921519 Review. Japanese.
Cited by
- PABPN1 suppresses TDP-43 toxicity in ALS disease models.
Chou CC, Alexeeva OM, Yamada S, Pribadi A, Zhang Y, Mo B, Williams KR, Zarnescu DC, Rossoll W. Chou CC, et al. Hum Mol Genet. 2015 Sep 15;24(18):5154-73. doi: 10.1093/hmg/ddv238. Epub 2015 Jun 30. Hum Mol Genet. 2015. PMID: 26130692 Free PMC article. - Identification of genetic modifiers of TDP-43 neurotoxicity in Drosophila.
Zhan L, Hanson KA, Kim SH, Tare A, Tibbetts RS. Zhan L, et al. PLoS One. 2013;8(2):e57214. doi: 10.1371/journal.pone.0057214. Epub 2013 Feb 27. PLoS One. 2013. PMID: 23468938 Free PMC article. - USP10 Inhibits Aberrant Cytoplasmic Aggregation of TDP-43 by Promoting Stress Granule Clearance.
Takahashi M, Kitaura H, Kakita A, Kakihana T, Katsuragi Y, Onodera O, Iwakura Y, Nawa H, Komatsu M, Fujii M. Takahashi M, et al. Mol Cell Biol. 2022 Mar 17;42(3):e0039321. doi: 10.1128/MCB.00393-21. Epub 2022 Jan 10. Mol Cell Biol. 2022. PMID: 35007165 Free PMC article. - Wild-type and A315T mutant TDP-43 exert differential neurotoxicity in a Drosophila model of ALS.
Estes PS, Boehringer A, Zwick R, Tang JE, Grigsby B, Zarnescu DC. Estes PS, et al. Hum Mol Genet. 2011 Jun 15;20(12):2308-21. doi: 10.1093/hmg/ddr124. Epub 2011 Mar 26. Hum Mol Genet. 2011. PMID: 21441568 Free PMC article. - RNA metabolism in neurodegenerative disease.
Liu EY, Cali CP, Lee EB. Liu EY, et al. Dis Model Mech. 2017 May 1;10(5):509-518. doi: 10.1242/dmm.028613. Dis Model Mech. 2017. PMID: 28468937 Free PMC article. Review.
References
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. - PubMed
- Kabashi E, Valdmanis PN, Dion P, Spiegelman D, McConkey BJ, et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet. 2008;40:572–574. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous