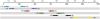Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions - PubMed (original) (raw)
Comparative Study
Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions
Marcus J Claesson et al. Nucleic Acids Res. 2010 Dec.
Abstract
High-throughput molecular technologies can profile microbial communities at high resolution even in complex environments like the intestinal microbiota. Recent improvements in next-generation sequencing technologies allow for even finer resolution. We compared phylogenetic profiling of both longer (454 Titanium) sequence reads with shorter, but more numerous, paired-end reads (Illumina). For both approaches, we targeted six tandem combinations of 16S rRNA gene variable regions, in microbial DNA extracted from a human faecal sample, in order to investigate their limitations and potentials. In silico evaluations predicted that the V3/V4 and V4/V5 regions would provide the highest classification accuracies for both technologies. However, experimental sequencing of the V3/V4 region revealed significant amplification bias compared to the other regions, emphasising the necessity for experimental validation of primer pairs. The latest developments of 454 and Illumina technologies offered higher resolution compared to their previous versions, and showed relative consistency with each other. However, the majority of the Illumina reads could not be classified down to genus level due to their shorter length and higher error rates beyond 60 nt. Nonetheless, with improved quality and longer reads, the far greater coverage of Illumina promises unparalleled insights into highly diverse and complex environments such as the human gut.
Figures
Figure 1.
Positions of primer sequences and tandem regions used in this work for 454 titanium and Illumina, mapped along 16S rRNA gene (co-ordinates based on the Escherichia coli 16S rRNA gene sequence). The arrows (∼100 bases) show approximately Illumina sequence read length.
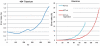
Figure 2.
Error rates as function of read lengths (provided by Roche and Fasteris). Error rates beyond 100 bp for Kit v4 were obtained through extrapolation [f(x) = 0.0763e0.0408_x_].
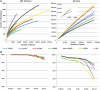
Figure 3.
Proportion of full-length 16S rRNA and tandem regions from simulated Titanium and Illumina reads, accurately classified at five taxonomic levels. Sequencing errors were also introduced using error rates above (dashed lines: KIT-v4; dotted lines: KIT-v3).
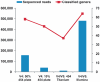
Figure 4.
Rarefaction curves for Titanium and Illumina reads at the 97% similarity phylotype level. Dashed lines are for 8277 randomly sub-sampled Titanium reads, equal in size to the smallest Titanium amplicon dataset. Illumina rarefaction curves were calculated from random sub-samplings of 229 048 reads, equal in size to the region with fewest reads (the V3/V4 region). The inset shows rarefaction curves from randomly sub-sampled Illumina reads equal in numbers to the corresponding 454 regions. (A) Proportion of sequenced Titanium and Illumina reads that were classified at four taxonomic levels (B) Single V4 reads sequenced in our earlier study (5) were included for comparison.
Figure 5.
Resolution at genus level for Titanium and Illumina V4/V5 reads. Single V4 reads sequenced at two different depths in our earlier study (5) were included for comparison.
Figure 6.
Relative phylum and genus abundances for sequence reads from both sequencing technologies. Single V4 reads sequenced using non-Titanium pyrosequencing in our earlier study (5) were included for comparison.
Figure 7.
MEGAN comparison based on BLAST searches of all 454 Titanium reads (A) and a random subset of Illumina reads (B) against an rRNA-specific database.
Similar articles
- Evaluation of 16S rRNA amplicon sequencing using two next-generation sequencing technologies for phylogenetic analysis of the rumen bacterial community in steers.
Myer PR, Kim M, Freetly HC, Smith TPL. Myer PR, et al. J Microbiol Methods. 2016 Aug;127:132-140. doi: 10.1016/j.mimet.2016.06.004. Epub 2016 Jun 6. J Microbiol Methods. 2016. PMID: 27282101 - Analysis of the intestinal microbiota using SOLiD 16S rRNA gene sequencing and SOLiD shotgun sequencing.
Mitra S, Förster-Fromme K, Damms-Machado A, Scheurenbrand T, Biskup S, Huson DH, Bischoff SC. Mitra S, et al. BMC Genomics. 2013;14 Suppl 5(Suppl 5):S16. doi: 10.1186/1471-2164-14-S5-S16. Epub 2013 Oct 16. BMC Genomics. 2013. PMID: 24564472 Free PMC article. - Optimisation of methods for bacterial skin microbiome investigation: primer selection and comparison of the 454 versus MiSeq platform.
Castelino M, Eyre S, Moat J, Fox G, Martin P, Ho P, Upton M, Barton A. Castelino M, et al. BMC Microbiol. 2017 Jan 21;17(1):23. doi: 10.1186/s12866-017-0927-4. BMC Microbiol. 2017. PMID: 28109256 Free PMC article. - Combining 16S rRNA gene variable regions enables high-resolution microbial community profiling.
Fuks G, Elgart M, Amir A, Zeisel A, Turnbaugh PJ, Soen Y, Shental N. Fuks G, et al. Microbiome. 2018 Jan 26;6(1):17. doi: 10.1186/s40168-017-0396-x. Microbiome. 2018. PMID: 29373999 Free PMC article. - An extended single-index multiplexed 16S rRNA sequencing for microbial community analysis on MiSeq illumina platforms.
Derakhshani H, Tun HM, Khafipour E. Derakhshani H, et al. J Basic Microbiol. 2016 Mar;56(3):321-6. doi: 10.1002/jobm.201500420. Epub 2015 Oct 1. J Basic Microbiol. 2016. PMID: 26426811
Cited by
- Variations in microbial community compositions and processes imposed under contrast geochemical contexts in Sicilian mud volcanoes, Italy.
Chen JN, Chiu YP, Tu TH, Italiano F, Wang PL, Lin LH. Chen JN, et al. Front Microbiol. 2024 Sep 20;15:1461252. doi: 10.3389/fmicb.2024.1461252. eCollection 2024. Front Microbiol. 2024. PMID: 39372275 Free PMC article. - Antibiotic subclasses differentially perturb the gut microbiota in kidney transplant recipients.
Dong H, Li R, Zhao N, Dadhania DM, Suthanthiran M, Lee JR, Ling W. Dong H, et al. Front Transplant. 2024 Sep 20;3:1400067. doi: 10.3389/frtra.2024.1400067. eCollection 2024. Front Transplant. 2024. PMID: 39371270 Free PMC article. - The effects of different lung parts, age, and batches on the lung microbiota of healthy rats.
Chen P, Hu T, Jiang H, Li B, Li G, Ran P, Zhou Y. Chen P, et al. Ann Med. 2024 Jul 16;56(1):2381085. doi: 10.1080/07853890.2024.2381085. Epub 2024 Aug 4. Ann Med. 2024. PMID: 39099020 Free PMC article. - Exploring the Impact of Short Term Travel on Gut Microbiota and Probiotic Bacteria Mediated Stability.
Zhao Y, Li C, Wu K, Chen H, Wang Q, Xiao Y, Yao S, Hong A, Zhang M, Lei S, Yang W, Zhong S, Umar A, Huang J, Yu Z. Zhao Y, et al. Biomedicines. 2024 Jun 21;12(7):1378. doi: 10.3390/biomedicines12071378. Biomedicines. 2024. PMID: 39061954 Free PMC article. - How the root bacterial community of Ficus tikoua responds to nematode infection: enrichments of nitrogen-fixing and nematode-antagonistic bacteria in the parasitized organs.
Meng XR, Gan Y, Liao LJ, Li CN, Wang R, Liu M, Deng JY, Chen Y. Meng XR, et al. Front Plant Sci. 2024 Jun 28;15:1374431. doi: 10.3389/fpls.2024.1374431. eCollection 2024. Front Plant Sci. 2024. PMID: 39006956 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials