Binding and cleavage of CRISPR RNA by Cas6 - PubMed (original) (raw)
Binding and cleavage of CRISPR RNA by Cas6
Jason Carte et al. RNA. 2010 Nov.
Abstract
The CRISPR-Cas system provides many prokaryotes with acquired resistance to viruses and other mobile genetic elements. The core components of this defense system are small, host-encoded prokaryotic silencing (psi)RNAs and Cas (CRISPR-associated) proteins. Invader-derived sequences within the psiRNAs guide Cas effector proteins to recognize and silence invader nucleic acids. Critical for CRISPR-Cas defense is processing of the psiRNAs from the primary transcripts of the host CRISPR (clustered regularly interspaced short palindromic repeat) locus. Cas6, a previously identified endoribonuclease present in a wide range of prokaryotes with the CRISPR-Cas system, binds and cleaves within the repeat sequences that separate the individual invader targeting elements in the CRISPR locus transcript. In the present study, we investigated several key aspects of the mechanism of function of Cas6 in psiRNA biogenesis. RNA footprinting reveals that Pyrococcus furiosus Cas6 binds to a 7-nt (nucleotide) sequence near the 5' end of the CRISPR RNA repeat sequence, 14 nt upstream of the Cas6 cleavage site. In addition, analysis of the cleavage activity of P. furiosus Cas6 proteins with mutations at conserved residues suggests that a triad comprised of Tyr31, His46, and Lys52 plays a critical role in catalysis, consistent with a possible general acid-base RNA cleavage mechanism for Cas6. Finally, we show that P. furiosus Cas6 remains stably associated with its cleavage products, suggesting additional roles for Cas6 in psiRNA biogenesis.
Figures
FIGURE 1.
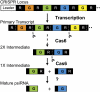
psiRNA biogenesis. The primary CRISPR locus transcript contains multiple guide sequences (color segments, G) separated by a repeat sequence (black segments, R). Cas6 cleaves within the repeats of a CRISPR transcript (cleavage site labeled with dotted line), eventually producing individual RNAs comprised of an 8-nt repeat sequence (i.e., psi-tag), ∼37-nt guide sequence, and 22-nt repeat sequence (1× intermediate). These “1× intermediate RNAs” are the end-products of Cas6 cleavage. The 22-nt repeat sequence found at the 3′ end of the 1× Cas6 product is subsequently removed from the mature psiRNAs (that guide the Cmr complex to cleave target RNAs in P. furiosus) by an unknown activity. Cas6 cleavage proceeds through a series of intermediates including the 2× intermediate shown here.
FIGURE 2.
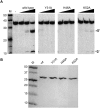
Lead-induced and RNase A cleavage protection of CRISPR repeat RNA by P. furiosus Cas6. (A) 3′-end-labeled P. furiosus CRISPR repeat RNA was incubated in the absence (RNA) or presence of increasing concentrations of Cas6 (indicated as micromolar, μM) and subjected to RNase A cleavage (left panel) or lead-induced cleavage (right panel). RNAs were separated on 15% denaturing (7 M urea) polyacrylamide gels. Size markers include 5′-end-labeled RNA markers (M) and alkaline hydrolysis ladders (OH). (Blue bars) Sites of strong protection. (B) Cleavage protection assays performed as in A with 5′-end-labeled CRISPR repeat RNA. A summary of cleavage protections is displayed to the right of each gel. (C) A summary of observed cleavage protection is shown. The Cas6 cleavage site is indicated by an asterisk (*).
FIGURE 3.
Cleavage activity of Cas6 mutants. (A) Uniformly 32P-labeled CRISPR repeat RNA was incubated in the absence (−) or presence of increasing concentrations of wild-type or mutant Cas6 (10, 50, and 500 nM) followed by separation on a 15% denaturing (7 M urea) polyacrylamide gel. The 5′ and 3′ cleavage products are indicated to the right. (B) Purified wild-type (wt) and mutant Cas6 proteins were analyzed by SDS-PAGE and Coomassie blue staining. Molecular weight markers are indicated in kilodaltons.
FIGURE 4.
Substrate binding activity of Cas6 mutants. Uniformly 32P-labeled CRISPR repeat RNA was incubated in the absence (−) or presence of increasing concentrations of wild-type or mutant Cas6 (1, 50, 200, and 500 nM). Formation of stable complexes was assessed by native gel mobility shift analysis. The positions of the free (RNA) and bound (RNP) substrate RNA are indicated.
FIGURE 5.
Immunopurified Cas6 cleaves CRISPR repeat RNA and associates with substrate and product RNAs. (A) Cleavage activity. Uniformly 32P-labeled CRISPR repeat RNA was incubated in the absence (RNA) or presence of recombinant Cas6 (rCas6), whole cell extract (WCE), or samples from immunoprecipitation reactions using Cas6 antibodies; (Pre) preimmune; (Imm) immune; (S) supernatant; (P) pellet. The RNAs were separated on a 15% denaturing, 7 M urea-containing polyacrylamide gel, along with 5′-end-labeled RNA markers (M). (B) Northern blot analysis of Cas6 immunoprecipitation. RNAs extracted from WCE or from IPs using preimmune (Pre) or immune (Imm) Cas6 or Cmr2 antibodies were separated on 15% denaturing, 7 M urea-containing polyacrylamide gels, along with 5′-end-labeled RNA markers (M). A 5′-end-labeled DNA oligonucleotide antisense to psiRNA 6.01-specific sequences, or to the repeat sequence of P. furiosus CRISPRs 1, 5, and 6 were used as probes, as indicated. The positions of the 2× intermediate (2×), 1× intermediate (1×), and mature psiRNAs (arrows) are indicated.
Similar articles
- Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes.
Carte J, Wang R, Li H, Terns RM, Terns MP. Carte J, et al. Genes Dev. 2008 Dec 15;22(24):3489-96. doi: 10.1101/gad.1742908. Genes Dev. 2008. PMID: 19141480 Free PMC article. - Prokaryotic silencing (psi)RNAs in Pyrococcus furiosus.
Hale C, Kleppe K, Terns RM, Terns MP. Hale C, et al. RNA. 2008 Dec;14(12):2572-9. doi: 10.1261/rna.1246808. Epub 2008 Oct 29. RNA. 2008. PMID: 18971321 Free PMC article. - Interaction of the Cas6 riboendonuclease with CRISPR RNAs: recognition and cleavage.
Wang R, Preamplume G, Terns MP, Terns RM, Li H. Wang R, et al. Structure. 2011 Feb 9;19(2):257-64. doi: 10.1016/j.str.2010.11.014. Structure. 2011. PMID: 21300293 Free PMC article. - The RNA- and DNA-targeting CRISPR-Cas immune systems of Pyrococcus furiosus.
Terns RM, Terns MP. Terns RM, et al. Biochem Soc Trans. 2013 Dec;41(6):1416-21. doi: 10.1042/BST20130056. Biochem Soc Trans. 2013. PMID: 24256230 Free PMC article. Review. - Cutting it close: CRISPR-associated endoribonuclease structure and function.
Hochstrasser ML, Doudna JA. Hochstrasser ML, et al. Trends Biochem Sci. 2015 Jan;40(1):58-66. doi: 10.1016/j.tibs.2014.10.007. Epub 2014 Nov 18. Trends Biochem Sci. 2015. PMID: 25468820 Review.
Cited by
- Function and regulation of clustered regularly interspaced short palindromic repeats (CRISPR) / CRISPR associated (Cas) systems.
Richter C, Chang JT, Fineran PC. Richter C, et al. Viruses. 2012 Oct 19;4(10):2291-311. doi: 10.3390/v4102291. Viruses. 2012. PMID: 23202464 Free PMC article. Review. - Genome sequencing of a genetically tractable Pyrococcus furiosus strain reveals a highly dynamic genome.
Bridger SL, Lancaster WA, Poole FL 2nd, Schut GJ, Adams MW. Bridger SL, et al. J Bacteriol. 2012 Aug;194(15):4097-106. doi: 10.1128/JB.00439-12. Epub 2012 May 25. J Bacteriol. 2012. PMID: 22636780 Free PMC article. - Modulating the Cascade architecture of a minimal Type I-F CRISPR-Cas system.
Gleditzsch D, Müller-Esparza H, Pausch P, Sharma K, Dwarakanath S, Urlaub H, Bange G, Randau L. Gleditzsch D, et al. Nucleic Acids Res. 2016 Jul 8;44(12):5872-82. doi: 10.1093/nar/gkw469. Epub 2016 May 23. Nucleic Acids Res. 2016. PMID: 27216815 Free PMC article. - CRISPR interference (CRISPRi) for sequence-specific control of gene expression.
Larson MH, Gilbert LA, Wang X, Lim WA, Weissman JS, Qi LS. Larson MH, et al. Nat Protoc. 2013 Nov;8(11):2180-96. doi: 10.1038/nprot.2013.132. Epub 2013 Oct 17. Nat Protoc. 2013. PMID: 24136345 Free PMC article. - Structural basis of Type IV CRISPR RNA biogenesis by a Cas6 endoribonuclease.
Taylor HN, Warner EE, Armbrust MJ, Crowley VM, Olsen KJ, Jackson RN. Taylor HN, et al. RNA Biol. 2019 Oct;16(10):1438-1447. doi: 10.1080/15476286.2019.1634965. Epub 2019 Jun 28. RNA Biol. 2019. PMID: 31232162 Free PMC article.
References
- Andersson AF, Banfield JF 2008. Virus population dynamics and acquired virus resistance in natural microbial communities. Science 320: 1047–1050 - PubMed
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315: 1709–1712 - PubMed
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD 2005. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151: 2551–2561 - PubMed
- Brunel C, Romby P 2000. Probing RNA structure and RNA–ligand complexes with chemical probes. Methods Enzymol 318: 3–21 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials