The differential activity of interferon-α subtypes is consistent among distinct target genes and cell types - PubMed (original) (raw)
The differential activity of interferon-α subtypes is consistent among distinct target genes and cell types
Herwig P Moll et al. Cytokine. 2011 Jan.
Abstract
IFN-α proteins have been described to originate from 14 individual genes and allelic variants. However, the exceptional diversity of IFN-α and its functional impact are still poorly understood. To characterize the biological activity of IFN-α subtypes in relation to the cellular background, we investigated the effect of IFN-α treatment in primary fibroblasts and endothelial cells of vascular or lymphatic origin. The cellular response was evaluated for 13 distinct IFN-α proteins with respect to transcript regulation of the IFN-stimulated genes (ISGs) IFIT1, ISG15, CXCL10, CXCL11 and CCL8. The IFN-α proteins displayed a remarkably consistent potency in gene induction irrespective of target gene and cellular background which led to the classification of IFN-α subtypes with low (IFN-α1), intermediate (IFN-α2a, -4a, -4b, -5, -16, -21) and high (IFN-α2b, -6, -7, -8, -10, -14) activity. The differential potency of IFN-α classes was confirmed at the ISG protein level and the functional protection of cells against influenza virus infection. Differences in IFN activity were only observed at subsaturating levels of IFN-α proteins and did not affect the time course of ISG regulation. Cell-type specific responses were apparent for distinct target genes independent of IFN-α subtype and were based on different levels of basal versus inducible gene expression. While fibroblasts presented with a high constitutive level of IFIT1, the expression in endothelial cells was strongly induced by IFN-α. In contrast, CXCL10 and CXCL11 transcript levels were generally higher in endothelial cells despite a pronounced induction by IFN-α in fibroblasts. In summary, the divergent potency of IFN-α proteins and the cell-type specific regulation of individual IFN target genes may allow for the fine tuning of cellular responses to pathogen defense.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures
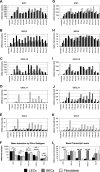
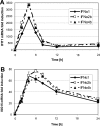
Fig. 1
ISG mRNA induction and transcript levels upon IFN-α stimulation of LECs, BECs and fibroblasts. Primary cells were stimulated with 100 pg/ml of each IFN-α subtype. Total RNA was analyzed by quantitative RT-PCR after 4 h of stimulation. The fold induction of IFIT1 (A), ISG15 (B), CXCL10 (C), CXCL11 (D), and CCL8 (E) mRNA was calculated in relation to the respective untreated control sample of each cell type. Three independent experiments were performed, the mean values of all experiments are given. Due to high differences in the overall induction values achieved in the three assays (biological variation) the standard deviations are not shown. (F) The mean induction value in response to all IFN-α subtypes was determined for the five target genes in the three distinct cell types. Mean and SD are given; significant differences (based on Student’s _T-_test) in ISG induction between cell types are indicated by one (p < 0.05) or two (p < 0.01) asterisks. (G–L) Since housekeeping gene expression (i.e. transcript level in relation to total RNA) did not vary substantially between LECs, BECs and fibroblasts (data not shown), ISG values were further normalized to a constant level of housekeeping gene transcripts to be able to compare ISG mRNA expression between cell types (“relative transcript level”). IFIT1 (G), ISG15 (H), CXCL10 (I), CXCL11 (J), and CCL8 (K) values were then expressed in relation to the untreated LEC sample (set to 1). (L) To illustrate the variance in basal ISG expression between cell types, ISG transcript levels of untreated cells were expressed in percent of the highest recorded value. Mean and SD of the three experiments are shown. Differences in basal ISG transcript levels between cell types were evaluated by Student’s _T_-test; significance levels of p < 0.05 and p < 0.01 are indicated by one and two asterisks, respectively.
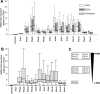
Fig. 2
Relative potency of IFN-α subtypes in ISG stimulation. Induction values for IFIT1, ISG15, CXCL10, CXCL11 and CCL8 were calculated in relation to IFN-α1 levels set to 1. Data distribution is illustrated by box plot and is given separately for LECs, BECs and fibroblasts (A) or for the combined set of data (B). The deduced classes of high, medium and low ISG inducers are summarized in (C).
Fig. 3
Comparison of ISG transcript and protein levels after IFN-α stimulation. LECs were treated with 100 pg/ml of IFN-α1, -2b, or -4b. At the indicated time points, cells and culture supernatant were harvested. Cell samples were subjected to concomitant mRNA and protein isolation. (A) IFIT1 and CXCL10 mRNA levels were analyzed by real-time RT-PCR. Data is given as mean and standard deviation of triplicate samples. (B) For analysis of IFIT1 protein expression, cytosolic extracts were subjected to SDS–PAGE and immunoblotting with anti-IFIT1 antiserum. For loading control, membranes were re-probed with anti-GAPDH antibody. (C) Culture supernatant was analyzed for CXCL10 protein by ELISA.
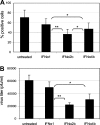
Fig. 4
Antiviral activity of IFN-α subtypes. LECs were exposed to IFN-α for 16 h or were left untreated before infection with influenza A (PR8 wt) virus at an MOI equaling 1. (A) Cells positive for intracellular expression of viral matrix and nucleoprotein were determined 7 h post infection by flow cytometry (n = 6). (B) The amount of virus released into the supernatant 24 h after virus infection was determined by a standard plaque formation assay (n = 4). Data presented are the mean and SD of all experiments performed. Differences in anti-viral activity of IFN-α subtypes were evaluated by Student’s _T_-test; significance levels of p < 0.05 and p < 0.01 are indicated by one and two asterisks, respectively.
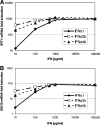
Fig. 5
Dose dependence of ISG regulation by IFN-α subtypes. LECs were treated with increasing concentrations of IFN-α1, -2b, or -4b for 4 h. Transcript levels of IFIT1 (A) and ISG15 (B) were analyzed by real-time RT-PCR. Data is given as mean and standard deviation of triplicate samples.
Fig. 6
Kinetics of ISG induction in response to IFN-α subtypes. LECs were exposed to 400 pg/ml of IFN-α1, 200 pg/ml of IFN-α4b, or 40 pg/ml of IFN-α2b for 2, 4, 6, 8, 12 and 24 h. Induction of IFIT1 (A) and ISG15 (B) mRNA was analyzed by real-time RT-PCR. Data is given as mean and standard deviation of triplicate samples.
Similar articles
- Dose-Dependent Differences in HIV Inhibition by Different Interferon Alpha Subtypes While Having Overall Similar Biologic Effects.
Schlaepfer E, Fahrny A, Gruenbach M, Kuster SP, Simon V, Schreiber G, Speck RF. Schlaepfer E, et al. mSphere. 2019 Feb 13;4(1):e00637-18. doi: 10.1128/mSphere.00637-18. mSphere. 2019. PMID: 30760614 Free PMC article. - Differential responses to IFN-alpha subtypes in human T cells and dendritic cells.
Hilkens CM, Schlaak JF, Kerr IM. Hilkens CM, et al. J Immunol. 2003 Nov 15;171(10):5255-63. doi: 10.4049/jimmunol.171.10.5255. J Immunol. 2003. PMID: 14607926 - Interferon-gamma potentiates the antiviral activity and the expression of interferon-stimulated genes induced by interferon-alpha in U937 cells.
Improta T, Pine R, Pfeffer LM. Improta T, et al. J Interferon Res. 1992 Apr;12(2):87-94. doi: 10.1089/jir.1992.12.87. J Interferon Res. 1992. PMID: 1315834 - Interferon-induced gene expression is a stronger predictor of treatment response than IL28B genotype in patients with hepatitis C.
Dill MT, Duong FH, Vogt JE, Bibert S, Bochud PY, Terracciano L, Papassotiropoulos A, Roth V, Heim MH. Dill MT, et al. Gastroenterology. 2011 Mar;140(3):1021-31. doi: 10.1053/j.gastro.2010.11.039. Epub 2010 Nov 25. Gastroenterology. 2011. PMID: 21111740
Cited by
- Antiviral potential of human IFN-α subtypes against influenza A H3N2 infection in human lung explants reveals subtype-specific activities.
Matos ADR, Wunderlich K, Schloer S, Schughart K, Geffers R, Seders M, Witt M, Christersson A, Wiewrodt R, Wiebe K, Barth P, Hocke A, Hippenstiel S, Hönzke K, Dittmer U, Sutter K, Rescher U, Rodionycheva S, Matera N, Ludwig S, Brunotte L. Matos ADR, et al. Emerg Microbes Infect. 2019;8(1):1763-1776. doi: 10.1080/22221751.2019.1698271. Emerg Microbes Infect. 2019. PMID: 31826721 Free PMC article. - Dose-Dependent Differences in HIV Inhibition by Different Interferon Alpha Subtypes While Having Overall Similar Biologic Effects.
Schlaepfer E, Fahrny A, Gruenbach M, Kuster SP, Simon V, Schreiber G, Speck RF. Schlaepfer E, et al. mSphere. 2019 Feb 13;4(1):e00637-18. doi: 10.1128/mSphere.00637-18. mSphere. 2019. PMID: 30760614 Free PMC article. - The Role of Cutaneous Type I IFNs in Autoimmune and Autoinflammatory Diseases.
Turnier JL, Kahlenberg JM. Turnier JL, et al. J Immunol. 2020 Dec 1;205(11):2941-2950. doi: 10.4049/jimmunol.2000596. J Immunol. 2020. PMID: 33229366 Free PMC article. Review. - Interferon-Regulated Expression of Cellular Splicing Factors Modulates Multiple Levels of HIV-1 Gene Expression and Replication.
Roesmann F, Müller L, Klaassen K, Heß S, Widera M. Roesmann F, et al. Viruses. 2024 Jun 11;16(6):938. doi: 10.3390/v16060938. Viruses. 2024. PMID: 38932230 Free PMC article. Review. - Quadruple therapy for asymptomatic COVID-19 infection patients.
Wang L, Xu X, Ruan J, Lin S, Jiang J, Ye H. Wang L, et al. Expert Rev Anti Infect Ther. 2020 Jul;18(7):617-624. doi: 10.1080/14787210.2020.1758066. Epub 2020 May 3. Expert Rev Anti Infect Ther. 2020. PMID: 32362193 Free PMC article.
References
- Isaacs A., Lindenmann J. Virus interference. I. The interferon. Proc R Soc Lond B Biol Sci. 1957;147:258–267. - PubMed
- Pestka S., Krause C.D., Walter M.R. Interferons, interferon-like cytokines, and their receptors. Immunol Rev. 2004;202:8–32. - PubMed
- Diaz M.O., Pomykala H.M., Bohlander S.K., Maltepe E., Malik K., Brownstein B. Structure of the human type-I interferon gene cluster determined from a YAC clone contig. Genomics. 1994;22:540–552. - PubMed
- Chen J., Baig E., Fish E.N. Diversity and relatedness among the type I interferons. J Interferon Cytokine Res. 2004;24:687–698. - PubMed
- Platanias L.C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–386. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous