The nucleolus under stress - PubMed (original) (raw)
Review
The nucleolus under stress
Séverine Boulon et al. Mol Cell. 2010.
Abstract
Cells typically respond quickly to stress, altering their metabolism to compensate. In mammalian cells, stress signaling usually leads to either cell-cycle arrest or apoptosis, depending on the severity of the insult and the ability of the cell to recover. Stress also often leads to reorganization of nuclear architecture, reflecting the simultaneous inhibition of major nuclear pathways (e.g., replication and transcription) and activation of specific stress responses (e.g., DNA repair). In this review, we focus on how two nuclear organelles, the nucleolus and the Cajal body, respond to stress. The nucleolus senses stress and is a central hub for coordinating the stress response. We review nucleolar function in the stress-induced regulation of p53 and the specific changes in nucleolar morphology and composition that occur upon stress. Crosstalk between nucleoli and CBs is also discussed in the context of stress responses.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures
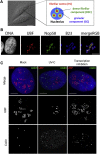
Figure 1
Overview of Nucleolar Organization under Physiological Conditions in the Mammalian Cell Nucleus, and Visualization by Immunofluorescence of Stress-Induced Changes to Nucleolar and Cajal Body Organization (A) Differential interference contrast (DIC) image of live HeLa cells: nucleoli are readily observed as phase-dense structures. Scale bar, 15 μm (left panel). Schematic representation of nucleolar tripartite internal organization, formed by the fibrillar center (FC), the dense fibrillar component (DFC), and the granular component (GC) (right panel). (B) Fluorescence microscopy images showing the three subnucleolar compartments in human U2OS cells. FC is visualized using antibodies against UBF, DFC using antibodies against Nop58, and GC using antibodies against B23/NPM. Scale bar, 10 μm. (C) Examples of stress-induced changes in nucleolar and CB organization in U2OS cells. (Left panel) Untreated cells. (Middle panel) UV-C-treated cells (6 hr postirradiation, 30 J/m2). (Right panel) DRB-treated cells (3 hr, 25 μg/mL). All images show UBF (nucleolar fibrillar center) in red and coilin (CB) in green. UV-C treatment induces nucleolar segregation and relocalization of coilin to nucleoplasmic microfoci. In contrast, DRB treatment induces nucleolar fragmentation and unravelling of the FC, as well as CB disruption and association of coilin with the nucleolus in cap-like structures. Scale bar, 5 μm.
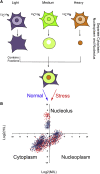
Figure 2
“Spatial Proteomics” Approach to Study Nucleolar Proteome Dynamics in Response to Stress (A) Typical “spatial proteomics” protocol. Cells are labeled with heavy-isotope containing amino acids (SILAC-based labeling) prior to fractionation into cytoplasm, nucleoplasm, and nucleolus. A whole-cell extract is created by recombining differentially labeled subcellular fractions and analyzed by LC-MS/MS. This can be used to measure the relative distribution of cellular proteins between the three compartments (medium/light [M/L] ratio, nucleoplasm/cytoplasm; heavy/medium [H/L] ratio, nucleolus/cytoplasm ratio) and analyze the stress-induced changes in protein localization, as visualized in (B).
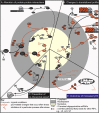
Figure 3
Overview of the Nucleolar-Related, Stress-Induced Mechanisms that Result in Increased p53 Activity The different mechanisms have been separated into three broad categories, namely those that primarily involve either (A) alterations of protein-protein interactions, (B) changes to the translational profile, or (C) prevention of coribosomal export of p53 and Hdmd2. The size of each quadrant represents their relative importance according to current literature. Grayscale text and objects indicate processes that occur during normal physiological conditions, whereas orange text and objects show the changes that occur following stress that result in increased p53 levels and activity. The outcomes of increases p53 are summarized in (D), namely the upregulation of p53 target genes which ultimately leads to either cell-cycle arrest or apoptosis, and the inhibition of ribosome biogenesis via the inhibition of rRNA transcription. Refer to text for detailed explanations of these pathways and for references.
Similar articles
- Reactive nucleolar and Cajal body responses to proteasome inhibition in sensory ganglion neurons.
Palanca A, Casafont I, Berciano MT, Lafarga M. Palanca A, et al. Biochim Biophys Acta. 2014 Jun;1842(6):848-59. doi: 10.1016/j.bbadis.2013.11.016. Epub 2013 Nov 19. Biochim Biophys Acta. 2014. PMID: 24269586 - Localization of α-Dystrobrevin in Cajal Bodies and Nucleoli: A New Role for α-Dystrobrevin in the Structure/Stability of the Nucleolus.
Hernández-Ibarra JA, Laredo-Cisneros MS, Mondragón-González R, Santamaría-Guayasamín N, Cisneros B. Hernández-Ibarra JA, et al. J Cell Biochem. 2015 Dec;116(12):2755-65. doi: 10.1002/jcb.25218. J Cell Biochem. 2015. PMID: 25959029 - Nuclear bodies and compartments: functional roles and cellular signalling in health and disease.
Zimber A, Nguyen QD, Gespach C. Zimber A, et al. Cell Signal. 2004 Oct;16(10):1085-104. doi: 10.1016/j.cellsig.2004.03.020. Cell Signal. 2004. PMID: 15240004 Review. - The Cajal body and the nucleolus: "In a relationship" or "It's complicated"?
Trinkle-Mulcahy L, Sleeman JE. Trinkle-Mulcahy L, et al. RNA Biol. 2017 Jun 3;14(6):739-751. doi: 10.1080/15476286.2016.1236169. Epub 2016 Sep 23. RNA Biol. 2017. PMID: 27661468 Free PMC article. Review. - [A role for Cajal bodies in assembly of the nuclear transcription machinery].
Gall JG. Gall JG. Tsitologiia. 2003;45(10):971-5. Tsitologiia. 2003. PMID: 14989168 Review. Russian.
Cited by
- METTL3/METTL14 maintain human nucleoli integrity by mediating SUV39H1/H2 degradation.
Shan Y, Zhang Y, Wei Y, Zhang C, Lin H, He J, Wang J, Guo W, Li H, Chen Q, Zhou T, Xing Q, Liu Y, Chen J, Pan G. Shan Y, et al. Nat Commun. 2024 Aug 21;15(1):7186. doi: 10.1038/s41467-024-51742-7. Nat Commun. 2024. PMID: 39169036 Free PMC article. - FTD/ALS-associated poly(GR) protein impairs the Notch pathway and is recruited by poly(GA) into cytoplasmic inclusions.
Yang D, Abdallah A, Li Z, Lu Y, Almeida S, Gao FB. Yang D, et al. Acta Neuropathol. 2015 Oct;130(4):525-35. doi: 10.1007/s00401-015-1448-6. Epub 2015 Jun 2. Acta Neuropathol. 2015. PMID: 26031661 Free PMC article. - A transcriptional program associated with cell cycle regulation predominates in the anti-inflammatory effects of CX-5461 in macrophage.
Wang J, Zheng Z, Cui X, Dai C, Li J, Zhang Q, Cheng M, Jiang F. Wang J, et al. Front Pharmacol. 2022 Oct 26;13:926317. doi: 10.3389/fphar.2022.926317. eCollection 2022. Front Pharmacol. 2022. PMID: 36386132 Free PMC article. - Epigenetic repression of ribosomal RNA transcription by ROCK-dependent aberrant cytoskeletal organization.
Wu TH, Kuo YY, Lee HH, Kuo JC, Ou MH, Chang ZF. Wu TH, et al. Sci Rep. 2016 Jun 28;6:28685. doi: 10.1038/srep28685. Sci Rep. 2016. PMID: 27350000 Free PMC article. - RNA-mediated pathogenic mechanisms in polyglutamine diseases and amyotrophic lateral sclerosis.
Chan HY. Chan HY. Front Cell Neurosci. 2014 Dec 19;8:431. doi: 10.3389/fncel.2014.00431. eCollection 2014. Front Cell Neurosci. 2014. PMID: 25565965 Free PMC article. Review.
References
- Al-Baker E.A., Oshin M., Hutchison C.J., Kill I.R. Analysis of UV-induced damage and repair in young and senescent human dermal fibroblasts using the comet assay. Mech. Ageing Dev. 2005;126:664–672. - PubMed
- Andersen J.S., Lam Y.W., Leung A.K., Ong S.E., Lyon C.E., Lamond A.I., Mann M. Nucleolar proteome dynamics. Nature. 2005;433:77–83. - PubMed
- Banerjee R., Weidman M.K., Navarro S., Comai L., Dasgupta A. Modifications of both selectivity factor and upstream binding factor contribute to poliovirus-mediated inhibition of RNA polymerase I transcription. J. Gen. Virol. 2005;86:2315–2322. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous