Epigenetic control of a VDR-governed feed-forward loop that regulates p21(waf1/cip1) expression and function in non-malignant prostate cells - PubMed (original) (raw)
. 2011 Mar;39(6):2045-56.
doi: 10.1093/nar/gkq875. Epub 2010 Nov 17.
Orla Maguire, Craig L Doig, Sebastiano Battaglia, Leah Fehr, Lara E Sucheston, Merja Heinaniemi, Laura P O'Neill, Christopher J McCabe, Bryan M Turner, Carsten Carlberg, Moray J Campbell
Affiliations
- PMID: 21088000
- PMCID: PMC3064804
- DOI: 10.1093/nar/gkq875
Epigenetic control of a VDR-governed feed-forward loop that regulates p21(waf1/cip1) expression and function in non-malignant prostate cells
James L Thorne et al. Nucleic Acids Res. 2011 Mar.
Abstract
In non-malignant RWPE-1 prostate epithelial cells signaling by the nuclear receptor Vitamin D Receptor (VDR, NR1I1) induces cell cycle arrest through targets including CDKN1A (encodes p21((waf1/cip1))). VDR dynamically induced individual histone modification patterns at three VDR binding sites (R1, 2, 3) on the CDKN1A promoter. The magnitude of these modifications was specific to each phase of the cell cycle. For example, H3K9ac enrichment occurred rapidly only at R2, whereas parallel accumulation of H3K27me3 occurred at R1; these events were significantly enriched in G(1) and S phase cells, respectively. The epigenetic events appeared to allow VDR actions to combine with p53 to enhance p21((waf1/cip1)) activation further. In parallel, VDR binding to the MCM7 gene induced H3K9ac enrichment associated with rapid mRNA up-regulation to generate miR-106b and consequently regulate p21((waf1/cip1)) expression. We conclude that VDR binding site- and promoter-specific patterns of histone modifications combine with miRNA co-regulation to form a VDR-regulated feed-forward loop to control p21((waf1/cip1)) expression and cell cycle arrest. Dissection of this feed-forward loop in a non-malignant prostate cell system illuminates mechanisms of sensitivity and therefore possible resistance in prostate and other VDR responsive cancers.
Figures
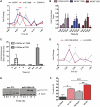
Figure 1.
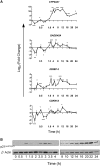
Dynamic regulation of VDR target genes. (A) RWPE-1 cells were treated with 1α,25(OH)2D3 (100 nM) or EtOH and mRNA was extracted at the indicated time points, and accumulation of indicated genes measured by TaqMan Q-RT–PCR. Accumulation of each target is given as log2 (fold change). Each data point represents the mean of triplicate experiments in triplicate wells ±SEM (*P < 0.05, **P < 0.01, ***P < 0.001). All measurements performed in technical and biological triplicate. (B) Total cell proteins were isolated from cells treated as above at the indicated time points (+D3) and subjected to western immunoblotting. Representative blots are shown for p21(waf1/cip1) (Abcam, ab7960). β-Actin used as a loading control (Abcam, ab8229). For quantification of signal Odyssey infrared imaging system (LI-COR, Lincoln, NE) was used and the quantification for the fold changes are under each image.
Figure 2.
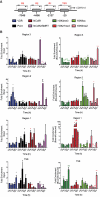
VDR-regulated epigenetic events on the promoter of CDKN1A. (A) The genomic location of the VDR binding regions (R3, R2 and R1) on CDKN1A and the transcription start site (TSS). (B) Left graphs. RWPE-1 bulk populations treated with 1α,25(OH)2D3 (100 nM) or EtOH for indicated time points. Association of RNA Pol II, VDR, NCOR1 and NCOR2/SMRT was measured at each region by X-ChIP using ChIP grade antibodies and normalized and given as fold enrichment over input as described previously (44). Primers are shown in Table 1. Enrichment was measured by Q-PCR with primers specific to these regions that amplified a product <150 bp. Right graphs. Parallel changes to histone modifications (H3K9me2, H3K27me3, H3K4me3 and H3K9ac) were assayed using N-ChIP and normalized using bound over unbound DNA pulled down with specific ChIP grade antibodies (42). N-ChIP was performed as previously described (53). Enrichment was measured by Q-PCR as above. All measurements performed in technical duplicate and biological triplicate.
Figure 3.
VDR interactions with co-repressors and the enhancement of 1α,25(OH)2D3-induced target gene expression. (A) RWPE-1 cells were treated as above, and X-ChIP performed and VDR DNA–protein complexes were eluted and immunoprecipitated with antibodies to NCOR1 or NCOR2/SMRT. Enrichment was measured by Q-PCR with primers specific to these regions that amplified a product less than 150 bp. Primers are shown in Table 1. All measurements performed in technical duplicate and biological triplicate. (B) RWPE-1 cells were treated with 1α,25(OH)2D3 (100 nM), 5-flurouracil (Fl-U) (50 nM), or combination (DF) for 1 h. Accumulation of CDKN1A and IGFBP3 measured by TaqMan Q-PCR. All measurements performed in technical and biological triplicate (*P < 0.05, **P < 0.01, ***P < 0.001).
Figure 4.
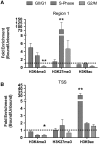
The cell cycle determines the magnitude of VDR-regulated histone modification. RWPE-1 cells were treated with 1α,25(OH)2D3 (100 nM) for 0.5 h prior to being FACS-sorted into the different phases of the cell cycle. Cells stained with Hoechst 33342 (Invitrogen), fractionated using a MoFlo cell sorter (Beckman-Coulter, High Wycombe, UK) and Summit V4.3 software (Beckman-Coulter, UK) and 5 × 105 cells/phase collected. Induced changes to repressive (H3K27me3) and activating (H3K4me3 and H3K9ac) histone modifications at R1 and the TSS on the CDKN1A promoter were interrogated using Carrier-ChIP protocols, C-ChIP was adapted from N-ChIP as previously described (46) by ‘spiking’ with SL2 cells prior to chromatin extraction and using human specific primers (Table 1). Histone modifications were assayed using N-ChIP and normalized using bound over unbound DNA pulled down with specific antibodies. Each data point represents the mean of triplicate experiments in duplicate wells ± S.E.M. (*P < 0.05, **P < 0.01).
Figure 5.
MiR-106b co-expression governs p21(waf1/cip1) expression and cell cycle arrest. (A) RWPE-1 cells were treated with 1α,25(OH)2D3 (100 nM) or EtOH control for indicated time points, mRNA extracted, and levels of accumulation of indicated genes measured by TaqMan Q-PCR. For miRNA quantitation, Q-PCR performed using Assay-on-Demand miR-106b and RNU48 probes. All measurements performed in technical and biological triplicate. (B) Cells were treated as in A, chromatin extracted and VDR binding to the indicated VDREs in the MCM7 promoter, and R2 on the CDKN1A promoter was measured, as indicated in Figure 2. (C) Changes to H3K9ac were assayed using N-ChIP and normalized using bound over unbound DNA pulled down with specific ChIP grade antibodies (42). N-ChIP was performed as previously described (53). Enrichment was measured by Q-PCR as above. All measurements performed in technical and biological triplicate. (D) 100nM si-miR-106b (MI0000734, Dharmacon) and scrambled constructs (IN-001005-01-05, Dharmacon) transiently transfected into cells. Following transfection, cells were treated as above. CDKN1A mRNA expression was measured by TaqMan Q-PCR as in Figure 1A. All data points on panels A and D represent the mean of triplicate experiments amplified in triplicate wells ± SEM (*P < 0.05, **P < 0.01, ***P < 0.005). (E) The effect of mir-106b knockdown on p21(waf1/cip1) expression. Cells were treated as in panel C and p21(waf1/cip1) detected by western blot as in Figure 1B, and both scr and siRNA blots exposed for 30 s. (F) The effect of 1α,25(OH)2D3 treatment on cell cycle arrest was measured in the indicated treatment groups. For FACS analysis, cells were stained with Propidium Iodide solution and run through a FACScan II (Becton Dickinson Biosciences). DNA histograms were analyzed using ModFit software (Verity Software House). Results were plotted as percent of cells seen in G1 phase and each data point represents the mean of three separate experiments ± SEM (*P < 0.05, **P < 0.01, ***P < 0.005).
Figure 6.
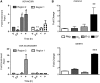
In vivo regulation of Cdkn1a and miR-106b in murine prostate. C57 BL/6xFVB WT mice were treated with 1α,25(OH)2D3 (20 μg/kg) or equal volume of EtOH for 12 or 24 h. Mice (n = 9) in each control and treatment group were sacrificed and the prostate removed and pooled into three groups of three, mRNA extracted using TRIzol (Invitrogen), and Cdkn1a and miR-106b levels were measured by TaqMan Q-PCR. Accumulation of each target is given as log2 (fold change). Each data point represents the mean ± SEM. A significant induction of miR-106b was observed at 12 h, and at 12 and 24 h Cdkn1a was significantly repressed. (**P < 0.01).
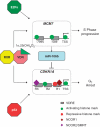
Figure 7.
A proposed VDR governed feed-forward loop to control p21(waf1/cip1). The VDR complex binds to both the CDKN1A gene [encodes p21(waf1/cip1)] and the DNA helicase gene MCM7. Over the first hour of activation only one response element on CDKN1A responds in a positive manner with loss of co-repressors and increased H3K9ac levels. The other response elements display repressive events. In parallel increased H3K9ac enrichment is observed at all response elements and the TSS of the MCM7 gene. The mixture of regulated epigenetic events on the CDKN1A promoter is significantly influenced by the status of the cell cycle and allows for cross-talk with p53, which also targets CDKN1A directly (response elements not shown) to sustain cell cycle arrest. The MCM7 gene is a target of S-phase transcription factors such as E2F family members and therefore this feed-forward loop fine tunes p21(waf1/cip1) expression in the context of cell cycle progression.
Similar articles
- Epigenetic distortion to VDR transcriptional regulation in prostate cancer cells.
Singh PK, Doig CL, Dhiman VK, Turner BM, Smiraglia DJ, Campbell MJ. Singh PK, et al. J Steroid Biochem Mol Biol. 2013 Jul;136:258-63. doi: 10.1016/j.jsbmb.2012.10.002. Epub 2012 Oct 23. J Steroid Biochem Mol Biol. 2013. PMID: 23098689 Free PMC article. Review. - Recruitment of NCOR1 to VDR target genes is enhanced in prostate cancer cells and associates with altered DNA methylation patterns.
Doig CL, Singh PK, Dhiman VK, Thorne JL, Battaglia S, Sobolewski M, Maguire O, O'Neill LP, Turner BM, McCabe CJ, Smiraglia DJ, Campbell MJ. Doig CL, et al. Carcinogenesis. 2013 Feb;34(2):248-56. doi: 10.1093/carcin/bgs331. Epub 2012 Oct 20. Carcinogenesis. 2013. PMID: 23087083 Free PMC article. - Regulation of the human p21(waf1/cip1) gene promoter via multiple binding sites for p53 and the vitamin D3 receptor.
Saramäki A, Banwell CM, Campbell MJ, Carlberg C. Saramäki A, et al. Nucleic Acids Res. 2006 Jan 24;34(2):543-54. doi: 10.1093/nar/gkj460. Print 2006. Nucleic Acids Res. 2006. PMID: 16434701 Free PMC article. - Elevated NCOR1 disrupts PPARalpha/gamma signaling in prostate cancer and forms a targetable epigenetic lesion.
Battaglia S, Maguire O, Thorne JL, Hornung LB, Doig CL, Liu S, Sucheston LE, Bianchi A, Khanim FL, Gommersall LM, Coulter HS, Rakha S, Giddings I, O'Neill LP, Cooper CS, McCabe CJ, Bunce CM, Campbell MJ. Battaglia S, et al. Carcinogenesis. 2010 Sep;31(9):1650-60. doi: 10.1093/carcin/bgq086. Epub 2010 May 13. Carcinogenesis. 2010. PMID: 20466759 Free PMC article. - Epigenetic corruption of VDR signalling in malignancy.
Abedin SA, Banwell CM, Colston KW, Carlberg C, Campbell MJ. Abedin SA, et al. Anticancer Res. 2006 Jul-Aug;26(4A):2557-66. Anticancer Res. 2006. PMID: 16886664 Review.
Cited by
- Mechanistic Effects of Calcitriol in Cancer Biology.
Díaz L, Díaz-Muñoz M, García-Gaytán AC, Méndez I. Díaz L, et al. Nutrients. 2015 Jun 19;7(6):5020-50. doi: 10.3390/nu7065020. Nutrients. 2015. PMID: 26102214 Free PMC article. Review. - H3K4me3 Is a Potential Mediator for Antiproliferative Effects of Calcitriol (1α,25(OH)2D3) in Ovarian Cancer Biology.
Han N, Jeschke U, Kuhn C, Hester A, Czogalla B, Mahner S, Rottmann M, Mayr D, Schmoeckel E, Trillsch F. Han N, et al. Int J Mol Sci. 2020 Mar 20;21(6):2151. doi: 10.3390/ijms21062151. Int J Mol Sci. 2020. PMID: 32245092 Free PMC article. - Vitamin d and susceptibility of chronic lung diseases: role of epigenetics.
Sundar IK, Rahman I. Sundar IK, et al. Front Pharmacol. 2011 Aug 30;2:50. doi: 10.3389/fphar.2011.00050. eCollection 2011. Front Pharmacol. 2011. PMID: 21941510 Free PMC article. - Epigenetic distortion to VDR transcriptional regulation in prostate cancer cells.
Singh PK, Doig CL, Dhiman VK, Turner BM, Smiraglia DJ, Campbell MJ. Singh PK, et al. J Steroid Biochem Mol Biol. 2013 Jul;136:258-63. doi: 10.1016/j.jsbmb.2012.10.002. Epub 2012 Oct 23. J Steroid Biochem Mol Biol. 2013. PMID: 23098689 Free PMC article. Review. - Integrated analysis of miRNA and mRNA during differentiation of human CD34+ cells delineates the regulatory roles of microRNA in hematopoiesis.
Raghavachari N, Liu P, Barb JJ, Yang Y, Wang R, Nguyen QT, Munson PJ. Raghavachari N, et al. Exp Hematol. 2014 Jan;42(1):14-27.e1-2. doi: 10.1016/j.exphem.2013.10.003. Epub 2013 Oct 16. Exp Hematol. 2014. PMID: 24139908 Free PMC article.
References
- Colston K, Colston MJ, Feldman D. 1,25-dihydroxyvitamin D3 and malignant melanoma: the presence of receptors and inhibition of cell growth in culture. Endocrinology. 1981;108:1083–1086. - PubMed
- Miyaura C, Abe E, Kuribayashi T, Tanaka H, Konno K, Nishii Y, Suda T. 1 alpha,25-Dihydroxyvitamin D3 induces differentiation of human myeloid leukemia cells. Biochem. Biophys. Res. Commun. 1981;102:937–943. - PubMed
- Campbell MJ, Elstner E, Holden S, Uskokovic M, Koeffler HP. Inhibition of proliferation of prostate cancer cells by a 19-nor-hexafluoride vitamin D3 analogue involves the induction of p21waf1, p27kip1 and E-cadherin. J. Mol. Endocrinol. 1997;19:15–27. - PubMed
- Elstner E, Campbell MJ, Munker R, Shintaku P, Binderup L, Heber D, Said J, Koeffler HP. Novel 20-epi-vitamin D3 analog combined with 9-cis-retinoic acid markedly inhibits colony growth of prostate cancer cells. Prostate. 1999;40:141–149. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01CA095367-06/CA/NCI NIH HHS/United States
- 2R01-CA-095045-06/CA/NCI NIH HHS/United States
- CA016056/CA/NCI NIH HHS/United States
- CRUK_/Cancer Research UK/United Kingdom
- C1015/A9077/PHS HHS/United States
- BB/D523651/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous