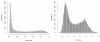Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis - PubMed (original) (raw)
Comparative Study
Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis
Pan Du et al. BMC Bioinformatics. 2010.
Abstract
Background: High-throughput profiling of DNA methylation status of CpG islands is crucial to understand the epigenetic regulation of genes. The microarray-based Infinium methylation assay by Illumina is one platform for low-cost high-throughput methylation profiling. Both Beta-value and M-value statistics have been used as metrics to measure methylation levels. However, there are no detailed studies of their relations and their strengths and limitations.
Results: We demonstrate that the relationship between the Beta-value and M-value methods is a Logit transformation, and show that the Beta-value method has severe heteroscedasticity for highly methylated or unmethylated CpG sites. In order to evaluate the performance of the Beta-value and M-value methods for identifying differentially methylated CpG sites, we designed a methylation titration experiment. The evaluation results show that the M-value method provides much better performance in terms of Detection Rate (DR) and True Positive Rate (TPR) for both highly methylated and unmethylated CpG sites. Imposing a minimum threshold of difference can improve the performance of the M-value method but not the Beta-value method. We also provide guidance for how to select the threshold of methylation differences.
Conclusions: The Beta-value has a more intuitive biological interpretation, but the M-value is more statistically valid for the differential analysis of methylation levels. Therefore, we recommend using the M-value method for conducting differential methylation analysis and including the Beta-value statistics when reporting the results to investigators.
Figures
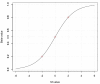
Figure 1
The relationship curve between M-value and Beta-value.
Figure 2
The histograms of Beta-value (left) and M-value (right) (27578 interrogated CpG sites in total).
Figure 3
The mean and standard deviation relations of technical replicates. Beta-value (left) and M-value (right).
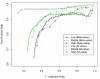
Figure 4
Performance comparisons of Beta- and M-value in the range of low, middle and high methylation levels based on the relationship of 1 - Detection Rate versus True Positive Rate.
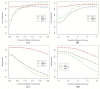
Figure 5
Performance comparisons of Beta and M-value based on the True Positive Rate (TPR) and Detection Rate (DR) at different thresholds of methylation difference. (A) TPR versus threshold of difference of Beta-value; (B) TPR versus threshold of difference of M-value; (C) DR versus threshold of difference of Beta-value; (D) DR versus threshold of difference of M-value.
Similar articles
- Purity estimation from differentially methylated sites using Illumina Infinium methylation microarray data.
Azim R, Wang S, Zhou S, Zhong X. Azim R, et al. Cell Cycle. 2020 Aug;19(16):2028-2039. doi: 10.1080/15384101.2020.1789315. Epub 2020 Jul 5. Cell Cycle. 2020. PMID: 32627651 Free PMC article. - Profiling DNA methylation differences between inbred mouse strains on the Illumina Human Infinium MethylationEPIC microarray.
Gujar H, Liang JW, Wong NC, Mozhui K. Gujar H, et al. PLoS One. 2018 Mar 12;13(3):e0193496. doi: 10.1371/journal.pone.0193496. eCollection 2018. PLoS One. 2018. PMID: 29529061 Free PMC article. - Using Illumina Infinium HumanMethylation 450K BeadChip to explore genome‑wide DNA methylation profiles in a human hepatocellular carcinoma cell line.
Sun N, Zhang J, Zhang C, Shi Y, Zhao B, Jiao A, Chen B. Sun N, et al. Mol Med Rep. 2018 Nov;18(5):4446-4456. doi: 10.3892/mmr.2018.9441. Epub 2018 Sep 3. Mol Med Rep. 2018. PMID: 30221710 Free PMC article. - Microarray-based method for detecting methylation changes of p16(Ink4a) gene 5'-CpG islands in gastric carcinomas.
Hou P, Shen JY, Ji MJ, He NY, Lu ZH. Hou P, et al. World J Gastroenterol. 2004 Dec 15;10(24):3553-8. doi: 10.3748/wjg.v10.i24.3553. World J Gastroenterol. 2004. PMID: 15534905 Free PMC article. - Monitoring methylation changes in cancer.
Beier V, Mund C, Hoheisel JD. Beier V, et al. Adv Biochem Eng Biotechnol. 2007;104:1-11. doi: 10.1007/10_024. Adv Biochem Eng Biotechnol. 2007. PMID: 17290816 Review.
Cited by
- Blood-based biomarkers of age-associated epigenetic changes in human islets associate with insulin secretion and diabetes.
Bacos K, Gillberg L, Volkov P, Olsson AH, Hansen T, Pedersen O, Gjesing AP, Eiberg H, Tuomi T, Almgren P, Groop L, Eliasson L, Vaag A, Dayeh T, Ling C. Bacos K, et al. Nat Commun. 2016 Mar 31;7:11089. doi: 10.1038/ncomms11089. Nat Commun. 2016. PMID: 27029739 Free PMC article. - Splicing-specific transcriptome-wide association uncovers genetic mechanisms for schizophrenia.
Hervoso JL, Amoah K, Dodson J, Choudhury M, Bhattacharya A, Quinones-Valdez G, Pasaniuc B, Xiao X. Hervoso JL, et al. Am J Hum Genet. 2024 Aug 8;111(8):1573-1587. doi: 10.1016/j.ajhg.2024.06.001. Epub 2024 Jun 25. Am J Hum Genet. 2024. PMID: 38925119 Free PMC article. - An Efficient Approach to Screening Epigenome-Wide Data.
Ray MA, Tong X, Lockett GA, Zhang H, Karmaus WJ. Ray MA, et al. Biomed Res Int. 2016;2016:2615348. doi: 10.1155/2016/2615348. Epub 2016 Mar 13. Biomed Res Int. 2016. PMID: 27034928 Free PMC article. - Epigenome-wide association study on asthma and chronic obstructive pulmonary disease overlap reveals aberrant DNA methylations related to clinical phenotypes.
Chen YC, Tsai YH, Wang CC, Liu SF, Chen TW, Fang WF, Lee CP, Hsu PY, Chao TY, Wu CC, Wei YF, Chang HC, Tsen CC, Chang YP, Lin MC; Taiwan Clinical Trial Consortium of Respiratory Disease (TCORE) group. Chen YC, et al. Sci Rep. 2021 Mar 3;11(1):5022. doi: 10.1038/s41598-021-83185-1. Sci Rep. 2021. PMID: 33658578 Free PMC article. - Purity estimation from differentially methylated sites using Illumina Infinium methylation microarray data.
Azim R, Wang S, Zhou S, Zhong X. Azim R, et al. Cell Cycle. 2020 Aug;19(16):2028-2039. doi: 10.1080/15384101.2020.1789315. Epub 2020 Jul 5. Cell Cycle. 2020. PMID: 32627651 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
- UL1 RR025741/RR/NCRR NIH HHS/United States
- UL1RR025741/RR/NCRR NIH HHS/United States
- 1RC1ES018461-01/ES/NIEHS NIH HHS/United States
- P30CA060553/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases