Histone3 lysine4 trimethylation regulated by the facilitates chromatin transcription complex is critical for DNA cleavage in class switch recombination - PubMed (original) (raw)
Histone3 lysine4 trimethylation regulated by the facilitates chromatin transcription complex is critical for DNA cleavage in class switch recombination
Andre Stanlie et al. Proc Natl Acad Sci U S A. 2010.
Abstract
Ig class switch recombination (CSR) requires expression of activation-induced cytidine deaminase (AID) and transcription through target switch (S) regions. Here we show that knockdown of the histone chaperone facilitates chromatin transcription (FACT) completely inhibited S region cleavage and CSR in IgA-switch-inducible CH12F3-2A B cells. FACT knockdown did not reduce AID or S region transcripts but did decrease histone3 lysine4 trimethylation (H3K4me3) at both the Sμ and Sα regions. Because knockdown of FACT or H3K4 methyltransferase cofactors inhibited DNA cleavage in H3K4me3-depleted S regions, H3K4me3 may serve as a mark for recruiting CSR recombinase. These findings revealed an unexpected evolutionary conservation between CSR and meiotic recombination.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
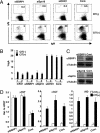
SSRP1 and Spt16 play a critical role in CSR. (A) Flow cytometry (FACS) profile of the IgA switching population following the introduction of the indicated RNAi oligonucleotide introduction under CIT(−) and (+) conditions. (B) Summarized data of IgA switching population of various gene knockdowns with respect to AID in CIT(−) and (+) conditions. Numbers 1 and 2 represent two independent RNAi oligonucleotides. SD values were determined from three independent experiments. (C) Knockdown efficiency of the indicated gene was quantified by immunobloting. (D) μGLT, αGLT, and AID transcripts were quantified by real-time PCR normalized to hypoxanthine-guanine phosphoribosyltransferase (HPRT) following the introduction of the indicated RNAi oligonucleotide. SD values were derived from three independent experiments.
Fig. 2.
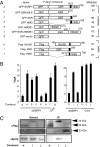
Rescue of CSR in SSRP1 knockdown cells expressing various SSRP1 deletion mutants and coimmunoprecipitation of SSRP1 and Spt16. (A) Representation of the various SSRP1 mutants and related constructs used in the CSR complementation experiment. Amino acid positions are indicated above each construct. All constructs used were well expressed (
Fig. S3
), and the expected molecular weights are shown on the right. NLS, nuclear localization signal. (B) Percentages of the IgA population as determined by FACS following the introduction of SSRP1 RNAi oligonucleotide along with various GFP- and Flag-tagged constructs. CSR efficiency was measured as the percentage of surface IgA expression in GFP-positive cells. SD values were determined from three independent experiments. (C) Immunoprecipitation (IP) by anti-Flag and the immunoblot analysis of Flag constructs and Spt16. Arrowheads indicate the expected molecular weights.
Fig. 3.
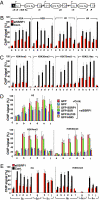
SSRP1 regulates the histone status of the S regions. (A) Schematic diagram of the position of the PCR products for ChIP assay. (B, C, and E) The ChIP assay was performed in SSRP1 knockdown and control samples using the indicated antibodies. No difference was observed between CIT(−) and (+) conditions. Data shown are under CIT(−) condition. (D) Rescue experiments of histone H3, H3K4me3 and H3K36me3 using various SSRP1 mutants in SSRP1 knockdown and control samples. Background values from controls with no antibody were subtracted (generally <5% of antibody signal). Values were normalized to the DNA input signals, followed by the maximum value in each data set. SD values were derived from three independent experiments.
Fig. 4.
H3K4me3 is critical for CSR. (A) Profile of the IgA switching population as determined by FACS in CH12F3-2A cells following the introduction of the indicated RNAi oligonucleotide. (B) Quantification of GLTs and AID transcripts by real-time PCR derived from the RNA of the indicated RNAi oligonucleotide samples normalized to HPRT. SD values were determined from three independent experiments. (C and D) (Top) Schematic diagram of the position of PCR products for the ChIP assay. The ChIP assay was performed in various knockdown and control samples using the indicated antibodies. Background values from controls with no antibody were subtracted (generally <5% of the antibody signal). Values were normalized to the DNA input signals followed by the maximum value in each data set. SD values were derived from three independent experiments.
Fig. 5.
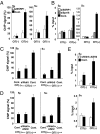
H3K4me3 is essential for S region DNA cleavage. (A) DNA break assay by γH2AX ChIP. The ChIP assay was performed in SSRP1 knockdown and control samples using an anti-γH2AX antibody. The pulled-down DNA was subjected to S- and Sα-specific detection by real-time PCR normalized to the fraction of γH2AX in H2AX, followed by the maximal value in each dataset. (B) The biotin-labeling break assay was performed in SSRP1 knockdown, Spt16 knockdown, and control samples. The pulled-down DNA was subjected to Sμ- and Sα specific detection by real-time PCR normalized to the input DNA. SD values were derived from three independent experiments. (C and D) The ChIP assay was performed in Wdr5 or Wdr5 and ASH2 knockdown and control samples using the anti-γH2AX antibody. (E) Biotin-labeling break assay was performed in Wdr5 and ASH2 knockdown and control samples.
Similar articles
- The histone chaperone Spt6 is required for activation-induced cytidine deaminase target determination through H3K4me3 regulation.
Begum NA, Stanlie A, Nakata M, Akiyama H, Honjo T. Begum NA, et al. J Biol Chem. 2012 Sep 21;287(39):32415-29. doi: 10.1074/jbc.M112.351569. Epub 2012 Jul 26. J Biol Chem. 2012. PMID: 22843687 Free PMC article. - Induction of activation-induced cytidine deaminase-targeting adaptor 14-3-3γ is mediated by NF-κB-dependent recruitment of CFP1 to the 5'-CpG-3'-rich 14-3-3γ promoter and is sustained by E2A.
Mai T, Pone EJ, Li G, Lam TS, Moehlman J, Xu Z, Casali P. Mai T, et al. J Immunol. 2013 Aug 15;191(4):1895-906. doi: 10.4049/jimmunol.1300922. Epub 2013 Jul 12. J Immunol. 2013. PMID: 23851690 Free PMC article. - The DSIF subunits Spt4 and Spt5 have distinct roles at various phases of immunoglobulin class switch recombination.
Stanlie A, Begum NA, Akiyama H, Honjo T. Stanlie A, et al. PLoS Genet. 2012;8(4):e1002675. doi: 10.1371/journal.pgen.1002675. Epub 2012 Apr 26. PLoS Genet. 2012. PMID: 22570620 Free PMC article. - Mechanism and control of V(D)J recombination versus class switch recombination: similarities and differences.
Dudley DD, Chaudhuri J, Bassing CH, Alt FW. Dudley DD, et al. Adv Immunol. 2005;86:43-112. doi: 10.1016/S0065-2776(04)86002-4. Adv Immunol. 2005. PMID: 15705419 Review. - Evolution of the immunoglobulin heavy chain class switch recombination mechanism.
Chaudhuri J, Basu U, Zarrin A, Yan C, Franco S, Perlot T, Vuong B, Wang J, Phan RT, Datta A, Manis J, Alt FW. Chaudhuri J, et al. Adv Immunol. 2007;94:157-214. doi: 10.1016/S0065-2776(06)94006-1. Adv Immunol. 2007. PMID: 17560275 Review.
Cited by
- Roles for histone H3K4 methyltransferase activities during immunoglobulin class-switch recombination.
Daniel JA, Nussenzweig A. Daniel JA, et al. Biochim Biophys Acta. 2012 Jul;1819(7):733-8. doi: 10.1016/j.bbagrm.2012.01.019. Epub 2012 Feb 12. Biochim Biophys Acta. 2012. PMID: 22710321 Free PMC article. Review. - Chromatin modifications and the DNA damage response to ionizing radiation.
Kumar R, Horikoshi N, Singh M, Gupta A, Misra HS, Albuquerque K, Hunt CR, Pandita TK. Kumar R, et al. Front Oncol. 2013 Jan 22;2:214. doi: 10.3389/fonc.2012.00214. eCollection 2012. Front Oncol. 2013. PMID: 23346550 Free PMC article. - Regulation of Aicda expression and AID activity.
Zan H, Casali P. Zan H, et al. Autoimmunity. 2013 Mar;46(2):83-101. doi: 10.3109/08916934.2012.749244. Epub 2013 Jan 17. Autoimmunity. 2013. PMID: 23181381 Free PMC article. Review. - The histone chaperone Spt6 is required for activation-induced cytidine deaminase target determination through H3K4me3 regulation.
Begum NA, Stanlie A, Nakata M, Akiyama H, Honjo T. Begum NA, et al. J Biol Chem. 2012 Sep 21;287(39):32415-29. doi: 10.1074/jbc.M112.351569. Epub 2012 Jul 26. J Biol Chem. 2012. PMID: 22843687 Free PMC article. - Induction of activation-induced cytidine deaminase-targeting adaptor 14-3-3γ is mediated by NF-κB-dependent recruitment of CFP1 to the 5'-CpG-3'-rich 14-3-3γ promoter and is sustained by E2A.
Mai T, Pone EJ, Li G, Lam TS, Moehlman J, Xu Z, Casali P. Mai T, et al. J Immunol. 2013 Aug 15;191(4):1895-906. doi: 10.4049/jimmunol.1300922. Epub 2013 Jul 12. J Immunol. 2013. PMID: 23851690 Free PMC article.
References
- Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. - PubMed
- Honjo T, Kinoshita K, Muramatsu M. Molecular mechanism of class switch recombination: Linkage with somatic hypermutation. Annu Rev Immunol. 2002;20:165–196. - PubMed
- Jung S, Rajewsky K, Radbruch A. Shutdown of class switch recombination by deletion of a switch region control element. Science. 1993;259:984–987. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous