Dioxygenases in Burkholderia ambifaria and Yersinia pestis that hydroxylate the outer Kdo unit of lipopolysaccharide - PubMed (original) (raw)
Dioxygenases in Burkholderia ambifaria and Yersinia pestis that hydroxylate the outer Kdo unit of lipopolysaccharide
Hak Suk Chung et al. Proc Natl Acad Sci U S A. 2011.
Abstract
Several gram-negative pathogens, including Yersinia pestis, Burkholderia cepacia, and Acinetobacter haemolyticus, synthesize an isosteric analog of 3-deoxy-D-manno-oct-2-ulosonic acid (Kdo), known as D-glycero-D-talo-oct-2-ulosonic acid (Ko), in which the axial hydrogen atom at the Kdo 3-position is replaced with OH. Here we report a unique Kdo 3-hydroxylase (KdoO) from Burkholderia ambifaria and Yersinia pestis, encoded by the bamb_0774 (BakdoO) and the y1812 (YpkdoO) genes, respectively. When expressed in heptosyl transferase-deficient Escherichia coli, these genes result in conversion of the outer Kdo unit of Kdo(2)-lipid A to Ko in an O(2)-dependent manner. KdoO contains the putative iron-binding motif, HXDX(n>40)H. Reconstitution of KdoO activity in vitro with Kdo(2)-lipid A as the substrate required addition of Fe(2+), α-ketoglutarate, and ascorbic acid, confirming that KdoO is a Fe(2+)/α-ketoglutarate/O(2)-dependent dioxygenase. Conversion of Kdo to Ko in Kdo(2)-lipid A conferred reduced susceptibility to mild acid hydrolysis. Although two enzymes that catalyze Fe(2+)/α-ketoglutarate/O(2)-dependent hydroxylation of deoxyuridine in fungal extracts have been reported previously, kdoO is the first example of a gene encoding a deoxy-sugar hydroxylase. Homologues of KdoO are found exclusively in gram-negative bacteria, including the human pathogens Burkholderia mallei, Yersinia pestis, Klebsiella pneumoniae, Legionella longbeachae, and Coxiella burnetii, as well as the plant pathogen Ralstonia solanacearum.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
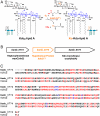
A proposed Kdo dioxygenase that generates the Ko moiety of B. ambifaria and Y. pestis LPS. A. A possible Fe2+/α-ketoglutarate/O2-dependent dioxygenase, designated KdoO, might account for the origin of the Ko unit after Kdo2-lipid A formation, consistent with the fact that Ko-containing bacteria make CMP-Kdo and transfer two Kdo residues to lipid A in the same manner as bacteria lacking Ko (15). LpxO, which generates the secondary _S_-2-hydroxymyristate substituent of Salmonella lipid A, provides a precedent for an analogous Kdo2-lipid A hydroxylation (17). B. The bamb_0774 gene, located between waaC and waaA on the B. ambifaria chromosome [Copeland A, et al. (2006) Complete sequence of chromosomes 1, 2, and 3 and plasmid 1 of Burkholderia cenocepacia AMMD, submitted August 2006 to the European Molecular Biology Laboratory/GenBank/DNA Data Base in Japan databases] (35), was identified as a possible structural gene for the Kdo hydroxylase because it encodes a protein with the active site signature of a Fe2+/α-ketoglutarate/O2-dependent dioxygenase (18). KdoO otherwise displays no sequence similarity to LpxO. C. Alignment of Bamb_0774 from B. ambifaria and Y1812 from Y. pestis KIM. These proteins share 52% identity (shown in red) and 64% similarity, and both contain the potential iron-binding motif, HXDX_n_H (n > 40) (potential downstream His residues shown in blue). Schematic representations: Kdo, black boxes; Ko, orange box; glucosamine, blue ovals; acyl chains, green squiggles; phosphate group, P.
Fig. 2.
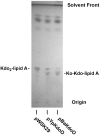
An altered form of Kdo2-lipid A in E. coli WBB06 expressing KdoO. Lipids were extracted (35) and separated by TLC, followed by charring with 10% sulfuric acid in ethanol. Cells harboring either pYpKdoO or pBaKdoO generated a slower migrating derivative of Kdo2-lipid A, consistent with Ko formation.
Fig. 3.
Kdo2-lipid A modification by KdoO involves hydroxylation and requires O2 A, B, and C. Cells expressing either YpKdoO or BaKdoO accumulate a Kdo2-lipid A derivative that is 16 amu larger than Kdo2-lipid A, consistent with the incorporation of a single oxygen atom and the generation of Ko-Kdo-lipid A (predicted [M-2H]2- at m/z 1,125.658). D and E. Growth in the absence of oxygen has no effect on the vector control but inhibits KdoO-dependent hydroxylation of Kdo2-lipid A. Schematic descriptions are the same as in Fig. 1.
Fig. 4.
18O from 18O2 is incorporated into the outer Kdo unit of Kdo2-lipid A by KdoO. A and B. KdoO expressing cells grown with 18O2 generate a Kdo2-lipid A derivative that is 1 m/z larger (as a doubly-charged species) than seen with 16O2-grown cells (blue vs. yellow spectra, respectively), confirming the incorporation of an O2-derived oxygen atom into the product. The asterisk* indicates the presence of the sodium adduct [M-3H + Na] ion of Kdo2-lipid A. C, D, and E. ESI-MS/MS analysis of Kdo2-lipid A, Ko-Kdo-lipid A, and [18O]Ko-Kdo-lipid A shows that YpKdoO modifies the outer Kdo unit, as evidenced in E by the [M-H]- ion of [18O]Ko at m/z 255.073, the [M-H216O-H]-ion of [18O]Ko at m/z 237.062, and the absence of corresponding [16O]Kdo fragments. Schematic descriptions are the same as in Fig. 1, except that [18O]Ko is indicated with a blue box.
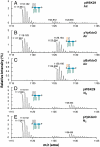
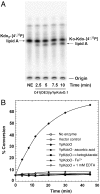
Fig. 5.
In vitro assay for KdoO and its cofactor requirements. A. Time course of the conversion of Kdo2-[4′-32P]lipid A to Ko-Kdo-[4′-32P]lipid A by membrane-free lysates (0.05 mg/mL) of C41(DE3)/pYpKdoO.1. Abbreviation: NE, no enzyme. B. KdoO activity was insert dependent and required added Fe2+, α-ketoglutarate, and ascorbic acid. KdoO activity was inhibited by EDTA. Reactions were initiated by adding washed membranes of C41(DE3)/pYpKdoO.1 (0.02 mg/mL final concentration) and terminated by spotting 2 μL portions of the reactions onto silica TLC plates at various times. The plates were developed in chloroform∶methanol∶acetic acid∶H2O (25∶15∶3.5∶4 vol/vol) and quantified with a PhosphorImager screen.
Similar articles
- Biochemical and Structural Insights into an Fe(II)/α-Ketoglutarate/O2-Dependent Dioxygenase, Kdo 3-Hydroxylase (KdoO).
Joo SH, Pemble CW 4th, Yang EG, Raetz CRH, Chung HS. Joo SH, et al. J Mol Biol. 2018 Oct 19;430(21):4036-4048. doi: 10.1016/j.jmb.2018.07.029. Epub 2018 Aug 7. J Mol Biol. 2018. PMID: 30092253 Free PMC article. - Kdo hydroxylase is an inner core assembly enzyme in the Ko-containing lipopolysaccharide biosynthesis.
Chung HS, Yang EG, Hwang D, Lee JE, Guan Z, Raetz CR. Chung HS, et al. Biochem Biophys Res Commun. 2014 Sep 26;452(3):789-94. doi: 10.1016/j.bbrc.2014.08.153. Epub 2014 Sep 6. Biochem Biophys Res Commun. 2014. PMID: 25204504 Free PMC article. - The core structure of the lipopolysaccharide from the causative agent of plague, Yersinia pestis.
Vinogradov EV, Lindner B, Kocharova NA, Senchenkova SN, Shashkov AS, Knirel YA, Holst O, Gremyakova TA, Shaikhutdinova RZ, Anisimov AP. Vinogradov EV, et al. Carbohydr Res. 2002 Apr 30;337(9):775-7. doi: 10.1016/s0008-6215(02)00074-5. Carbohydr Res. 2002. PMID: 11996830 - Biosynthesis of a novel 3-deoxy-D-manno-oct-2-ulosonic acid-containing outer core oligosaccharide in the lipopolysaccharide of Klebsiella pneumoniae.
Frirdich E, Vinogradov E, Whitfield C. Frirdich E, et al. J Biol Chem. 2004 Jul 2;279(27):27928-40. doi: 10.1074/jbc.M402549200. Epub 2004 Apr 15. J Biol Chem. 2004. PMID: 15090547
Cited by
- Lipopolysaccharide of the Yersinia pseudotuberculosis Complex.
Knirel YA, Anisimov AP, Kislichkina AA, Kondakova AN, Bystrova OV, Vagaiskaya AS, Shatalin KY, Shashkov AS, Dentovskaya SV. Knirel YA, et al. Biomolecules. 2021 Sep 26;11(10):1410. doi: 10.3390/biom11101410. Biomolecules. 2021. PMID: 34680043 Free PMC article. Review. - Variation, Modification and Engineering of Lipid A in Endotoxin of Gram-Negative Bacteria.
Kawahara K. Kawahara K. Int J Mol Sci. 2021 Feb 25;22(5):2281. doi: 10.3390/ijms22052281. Int J Mol Sci. 2021. PMID: 33668925 Free PMC article. Review. - Pushing the envelope: LPS modifications and their consequences.
Simpson BW, Trent MS. Simpson BW, et al. Nat Rev Microbiol. 2019 Jul;17(7):403-416. doi: 10.1038/s41579-019-0201-x. Nat Rev Microbiol. 2019. PMID: 31142822 Free PMC article. Review. - Biochemical and Structural Insights into an Fe(II)/α-Ketoglutarate/O2-Dependent Dioxygenase, Kdo 3-Hydroxylase (KdoO).
Joo SH, Pemble CW 4th, Yang EG, Raetz CRH, Chung HS. Joo SH, et al. J Mol Biol. 2018 Oct 19;430(21):4036-4048. doi: 10.1016/j.jmb.2018.07.029. Epub 2018 Aug 7. J Mol Biol. 2018. PMID: 30092253 Free PMC article. - Lipid A Has Significance for Optimal Growth of Coxiella burnetii in Macrophage-Like THP-1 Cells and to a Lesser Extent in Axenic Media and Non-phagocytic Cells.
Wang T, Yu Y, Liang X, Luo S, He Z, Sun Z, Jiang Y, Omsland A, Zhou P, Song L. Wang T, et al. Front Cell Infect Microbiol. 2018 Jun 8;8:192. doi: 10.3389/fcimb.2018.00192. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 29938202 Free PMC article.
References
- Miller SI, Ernst RK, Bader MW. LPS, TLR4 and infectious disease diversity. Nat Rev Microbiol. 2005;3:36–46. - PubMed
- Park BS, et al. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458:1191–1195. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 GM051796/GM/NIGMS NIH HHS/United States
- U54 GM069338/GM/NIGMS NIH HHS/United States
- R01 GM051796/GM/NIGMS NIH HHS/United States
- GM-51796/GM/NIGMS NIH HHS/United States
- GM-069338/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials