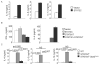The transcription factor STAT3 is required for T helper 2 cell development - PubMed (original) (raw)
The transcription factor STAT3 is required for T helper 2 cell development
Gretta L Stritesky et al. Immunity. 2011.
Abstract
Signal transducer and activator of transcription (STAT) family members direct the differentiation of T helper cells, with specific STAT proteins promoting distinct effector subsets. STAT6 is required for the development of T helper 2 (Th2) cells, whereas STAT3 promotes differentiation of Th17 and follicular helper T cell subsets. We demonstrated that STAT3 was also activated during Th2 cell development and was required for the expression of Th2 cell-associated cytokines and transcription factors. STAT3 bound directly to Th2 cell-associated gene loci and was required for the ability of STAT6 to bind target genes. In vivo, STAT3 deficiency in T cells eliminated the allergic inflammation in mice sensitized and challenged with ovalbumin or transgenic for constitutively active STAT6. Thus, STAT3 cooperates with STAT6 in promoting Th2 cell development. These results demonstrate that differentiating T helper cells integrate multiple STAT protein signals during Th2 cell development.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
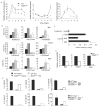
Figure 1. STAT3 is Activated During Th2 cell Differentiation
(A) Wild type (WT) and _Stat3Cd4_−/ − naïve CD4+ T cells were activated with anti-CD3 and anti-CD28 and cultured in IL-4 and anti-IFN-γ (Th2 conditions). Each day during differentiation, cells were stained for intracellular phospho-STAT3 and phospho-STAT6. Numbers in flow cytometry dot plots indicate the percentages of cells in each quadrant. (B) Graphical representation of percent phospo-STAT-positive cells and mean fluorescent intensity (mean ± s.d.). Data are an average of two mice and are representative of 2–3 experiments. (C) Naïve DO11.10 TCR transgenic T cells were stimulated with anti-CD3 or ovalbumin (OVA) in the presence of antigen-presenting cells and cultured under Th2 conditions. Each day during differentiation, cells were stained for intracellular phospho-STAT3 and phospho-STAT6. Numbers in flow cytometry dot plots indicate the percentages of cells in each quadrant. (D) Graphical representation of average percent phospho-STAT-positive cells for DO11.10 TCR transgenic T cells stimulated with OVA or anti-CD3 (mean ± s.d.). Data are an average of two mice and are representative of 2 experiments.
Figure 2. Reduced Th2 Cytokine Production in the Absence of STAT3
(A) WT naïve CD4+ T cells were activated with anti-CD3 and anti-CD28 and cultured under Th2 conditions, in the presence or absence of antibodies to the indicated cytokines and receptors (20 μg/ml per antibody) for five days before stimulation with anti-CD3. Twenty-four hours after stimulation supernatants were tested for cytokine concentration using ELISA (mean ± s.d.). Results are representative of experiments with four mice. (B) Cells cultured as in (A) with control antibody or the combination of antibodies indicated were tested for amount of pSTAT by intracellular staining on day 4 of Th2 culture. Numbers represent mean fluorescence intensity. Results are representative of experiments with four mice. (C) WT and _Stat3Cd4_−/ − naïve CD4+ T cells were activated with anti-CD3 and anti-CD28 and cultured with IL-4 + anti-IFN-γ (Th2 conditions). After 5 days of differentiation cells were collected and counted. Differentiated cells (1x106) were then re-stimulated with anti-CD3 (4 μg/ml) for 24 hours. Cell-free supernatant was collected and tested for various cytokines using ELISA. Data are the mean ± s.d. of results from two mice and representative of more than 5 experiments. Students t test was performed to calculate p values. (D) Cells activated and cultured as in A were re-stimulated with anti-CD3 for 5 hours. Following stimulation cells were stained with anti-IL-4 and anti-IL-9. Numbers in flow cytometry dot plots indicate the percentages of cells in the quadrant. (E) After re-stimulation of differentiated Th2 cells with anti-CD3 and recovery of supernatants as described in (A), cell pellets were collected, RNA was isolated, and quantitative PCR was performed for the indicated cytokines (mean ± s.d.). Data are representative of two experiments.
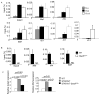
Figure 3. Reduced Th2-Specific Transcription Factor Expression in the Absence of STAT3
(A) WT and _Stat3Cd4_−/− naïve CD4+ T cells were differentiated under Th2 conditions and cells were isolated each day during differentiation. RNA isolated from cells was analyzed for gene expression using qPCR (mean ± s.d.). Data are representative of experiments with 4 mice. (B) WT and _Stat3Cd4_−/ − naïve CD4+ T cells were used directly or differentiated as in (A) for five days. Chromatin immunoprecipitation was performed for the indicated histone modifications and qPCR was used to determine the amounts of each modification (mean ± s.d.). (C) Chromatin immunoprecipitation from WT naïve CD4+ T cells with control or STAT3 antibodies followed by qPCR for detection of the indicated loci (mean ± s.d.). (D) WT and _Stat3Cd4_−/ − naïve CD4+ T cells were differentiated as in (A) for five days. Nuclei were isolated and left untreated or treated with micrococcal nuclease for 10 minutes. Relative accessibility is defined as 2 to the power of the difference between the Ct value of qPCR of untreated and treated samples (mean ± s.d.). (E) WT and _Stat3Cd4_−/ − naïve CD4+ T cells were differentiated as in (A). On Day 2 cells were transduced with control, Batf or _Maf_-expressing retrovirus. Sorted transduced cells were re-stimulated with anti-CD3 for 24 hours. Supernatant was tested for Th2 cytokines using ELISA (mean ± s.d.). Data are representative of 2 independent experiments.
Figure 4. STAT3 and STAT6 Cooperate in Promoting Th2 Cytokine Production
(A) Naïve CD4+ T cells were activated for 48 hours in the absence of skewing cytokines before being transduced with control or STAT3C-expressing retrovirus. After 5 days in culture, cells were sorted and re-stimulated with anti-CD3 (4 μg/ml) for 24 hours before cell-free supernatant was tested for cytokines using ELISA (mean ± s.d.). (B) Naïve CD4+ T cells were activated for 48 hours under Th1 conditions (5 ng/ml IL–12 + 10 μg/ml anti-IL-4) before being transduced with control, STAT6VT-expressing, STAT3-expressing or both retroviruses. After 5 days in culture, cells were sorted and re-stimulated with anti-CD3 (4 μg/ml) for 24 hours before cell-free supernatant was tested for cytokines using ELISA (mean ± s.d.). (C) CD4+ cells were isolated from WT, STAT6VT and STAT6VT-_Stat3Cd4_−/ − mice. Cells were then re-stimulated with anti-CD3 for 24 hours. Cell-free supernatant was collected and tested for various cytokines using ELISA. Data are representative of 2 experiments (average of 2–6 mice ± s.d.). Students t test was performed to calculate p values.
Figure 5. STAT3 Binds Th2-Associated Gene Loci and Defines the STAT6 Binding Pattern
(A) Naïve CD4+ T cells activated with anti-CD3 and anti-CD28 and cultured under Th1, Th2 or Th17 conditions for 3 days were used for ChIP performed with normal rabbit IgG or anti-STAT3 before qPCR was performed for the indicated genes. Data are expressed as percent input ± s.d. and control Ig background values are subtracted from the values indicated. Data are representative of 3–4 experiments with similar results. (B) Naïve CD4+ T cells from WT and _Stat3Cd4_−/ − activated with anti-CD3 and anti-CD28 and cultured under Th2 conditions for 3 days were used for ChIP performed with normal rabbit IgG or anti-STAT6 and qPCR was performed for the indicated genes. Data are expressed as percent input ± s.d. and control Ig background values are subtracted from the values indicated. Data are representative of 3–4 experiments with similar results. ND, not detected. (C) CD4+ cells were isolated from WT, STAT6VT and STAT6VT-_Stat3Cd4_−/ − mice. Cells were then re-stimulated with anti-CD3 for 6 hrs before RNA was isolated and quantitative PCR was performed. Data are representative of 2 experiments (average of 2–6 mice ± s.d.). Students t test was performed to calculate p values.
Figure 6. STAT3 Promotes the Development of Allergic Inflammation
(A–C) WT and _Stat3Cd4_−/ − mice were immunized with OVA and Alum on days 0 and 7 and challenged as described in methods. After challenges, BAL cell numbers were determined by cell counting and flow cytometry (A), and cytokine levels were measured in BAL fluid and in culture supernatants from splenocytes stimulated with OVA for 72 h using ELISA. Data are average of 5–6 mice per group ± s.e.m (D) Incidence of blepharitis and atopic dermatitis of WT, STAT6VT and STAT6VT-_Stat3Cd4_−/ − mice are shown. Incidence was determined by visual examination of mice (n=25 per group). (E) Numbers of eosinophils (defined by flow cytometry) recovered in BAL. BAL data are representative of 2 independent experiments and shown as the average of 2 mice per group ± s.d. For A–C and E, Students t test was performed to calculate p values. (F) Ear tissue from WT, STAT6VT and STAT6VT-_Stat3Cd4_−/ − mice were fixed and paraffin-embedded sections were stained with hematoxylin-eosin. Magnification is indicated in the panel and photomicrographs are representative of 10 mice per group (G) Lungs from WT, STAT6VT and STAT6VT-_Stat3Cd4_−/ − mice were embedded in paraffin and stained with H & E. Magnification is indicated in the panel and photomicrographs are representative of 10 mice per group.
Similar articles
- Poly-ADP-ribosyl polymerase-14 promotes T helper 17 and follicular T helper development.
Mehrotra P, Krishnamurthy P, Sun J, Goenka S, Kaplan MH. Mehrotra P, et al. Immunology. 2015 Dec;146(4):537-46. doi: 10.1111/imm.12515. Epub 2015 Sep 28. Immunology. 2015. PMID: 26222149 Free PMC article. - Signal transducer and activator of transcription 6 controls chemokine production and T helper cell type 2 cell trafficking in allergic pulmonary inflammation.
Mathew A, MacLean JA, DeHaan E, Tager AM, Green FH, Luster AD. Mathew A, et al. J Exp Med. 2001 May 7;193(9):1087-96. doi: 10.1084/jem.193.9.1087. J Exp Med. 2001. PMID: 11342593 Free PMC article. - STAT4 and STAT6, their role in cellular and humoral immunity and in diverse human diseases.
Tolomeo M, Cascio A. Tolomeo M, et al. Int Rev Immunol. 2024;43(6):394-418. doi: 10.1080/08830185.2024.2395274. Epub 2024 Aug 26. Int Rev Immunol. 2024. PMID: 39188021 Review. - The capacity of Th2 lymphocytes to deliver B-cell help requires expression of the transcription factor STAT3.
Mari N, Hercor M, Denanglaire S, Leo O, Andris F. Mari N, et al. Eur J Immunol. 2013 Jun;43(6):1489-98. doi: 10.1002/eji.201242938. Epub 2013 Apr 19. Eur J Immunol. 2013. PMID: 23504518 - Regulation of the T helper cell type 2 (Th2)/T regulatory cell (Treg) balance by IL-4 and STAT6.
Chapoval S, Dasgupta P, Dorsey NJ, Keegan AD. Chapoval S, et al. J Leukoc Biol. 2010 Jun;87(6):1011-8. doi: 10.1189/jlb.1209772. Epub 2010 Mar 24. J Leukoc Biol. 2010. PMID: 20335310 Free PMC article. Review.
Cited by
- Poly (ADP-ribose) polymerase 14 and its enzyme activity regulates T(H)2 differentiation and allergic airway disease.
Mehrotra P, Hollenbeck A, Riley JP, Li F, Patel RJ, Akhtar N, Goenka S. Mehrotra P, et al. J Allergy Clin Immunol. 2013 Feb;131(2):521-31.e1-12. doi: 10.1016/j.jaci.2012.06.015. Epub 2012 Jul 25. J Allergy Clin Immunol. 2013. PMID: 22841009 Free PMC article. - Influence of Aspergillus fumigatus conidia viability on murine pulmonary microRNA and mRNA expression following subchronic inhalation exposure.
Croston TL, Nayak AP, Lemons AR, Goldsmith WT, Gu JK, Germolec DR, Beezhold DH, Green BJ. Croston TL, et al. Clin Exp Allergy. 2016 Oct;46(10):1315-27. doi: 10.1111/cea.12783. Epub 2016 Sep 16. Clin Exp Allergy. 2016. PMID: 27473664 Free PMC article. - S100A4-neutralizing antibody suppresses spontaneous tumor progression, pre-metastatic niche formation and alters T-cell polarization balance.
Grum-Schwensen B, Klingelhöfer J, Beck M, Bonefeld CM, Hamerlik P, Guldberg P, Grigorian M, Lukanidin E, Ambartsumian N. Grum-Schwensen B, et al. BMC Cancer. 2015 Feb 12;15:44. doi: 10.1186/s12885-015-1034-2. BMC Cancer. 2015. PMID: 25884510 Free PMC article. - Scavenging reactive oxygen species selectively inhibits M2 macrophage polarization and their pro-tumorigenic function in part, via Stat3 suppression.
Griess B, Mir S, Datta K, Teoh-Fitzgerald M. Griess B, et al. Free Radic Biol Med. 2020 Feb 1;147:48-60. doi: 10.1016/j.freeradbiomed.2019.12.018. Epub 2019 Dec 19. Free Radic Biol Med. 2020. PMID: 31863907 Free PMC article. - The mediatory role of Majie cataplasm on inflammation of allergic asthma through transcription factors related to Th1 and Th2.
Ji W, Zhang Q, Shi H, Dong R, Ge D, Du X, Ren B, Wang X, Wang Q. Ji W, et al. Chin Med. 2020 May 24;15:53. doi: 10.1186/s13020-020-00334-w. eCollection 2020. Chin Med. 2020. PMID: 32489402 Free PMC article.
References
- Afkarian M, Sedy JR, Yang J, Jacobson NG, Cereb N, Yang SY, Murphy TL, Murphy KM. T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nat Immunol. 2002;3:549–557. - PubMed
- Akaishi H, Takeda K, Kaisho T, Shineha R, Satomi S, Takeda J, Akira S. Defective IL-2-mediated IL-2 receptor alpha chain expression in Stat3-deficient T lymphocytes. Int Immunol. 1998;10:1747–1751. - PubMed
- Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol. 2006;24:607–656. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 HL007910/HL/NHLBI NIH HHS/United States
- U19 AI070448/AI/NIAID NIH HHS/United States
- U19 AI070448-03/AI/NIAID NIH HHS/United States
- T32 AI060519/AI/NIAID NIH HHS/United States
- T32 HL07910/HL/NHLBI NIH HHS/United States
- U19 AI070448-04/AI/NIAID NIH HHS/United States
- U19 AI070448-05/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous