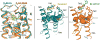Structure of a nanobody-stabilized active state of the β(2) adrenoceptor - PubMed (original) (raw)
. 2011 Jan 13;469(7329):175-80.
doi: 10.1038/nature09648.
Hee-Jung Choi, Juan Jose Fung, Els Pardon, Paola Casarosa, Pil Seok Chae, Brian T Devree, Daniel M Rosenbaum, Foon Sun Thian, Tong Sun Kobilka, Andreas Schnapp, Ingo Konetzki, Roger K Sunahara, Samuel H Gellman, Alexander Pautsch, Jan Steyaert, William I Weis, Brian K Kobilka
Affiliations
- PMID: 21228869
- PMCID: PMC3058308
- DOI: 10.1038/nature09648
Structure of a nanobody-stabilized active state of the β(2) adrenoceptor
Søren G F Rasmussen et al. Nature. 2011.
Abstract
G protein coupled receptors (GPCRs) exhibit a spectrum of functional behaviours in response to natural and synthetic ligands. Recent crystal structures provide insights into inactive states of several GPCRs. Efforts to obtain an agonist-bound active-state GPCR structure have proven difficult due to the inherent instability of this state in the absence of a G protein. We generated a camelid antibody fragment (nanobody) to the human β(2) adrenergic receptor (β(2)AR) that exhibits G protein-like behaviour, and obtained an agonist-bound, active-state crystal structure of the receptor-nanobody complex. Comparison with the inactive β(2)AR structure reveals subtle changes in the binding pocket; however, these small changes are associated with an 11 Å outward movement of the cytoplasmic end of transmembrane segment 6, and rearrangements of transmembrane segments 5 and 7 that are remarkably similar to those observed in opsin, an active form of rhodopsin. This structure provides insights into the process of agonist binding and activation.
Figures
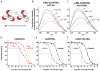
Figure 1. Effect of Nb80 on β2AR structure and function
a, The cartoon illustrates the movement of the environmentally-sensitive bimane probe attached to Cys2656.27 in the cytoplasmic end of TM6 from a more buried, hydrophobic environment to a more polar, solvent-exposed position during receptor activation that results in a decrease in the observed fluorescence in Figure 1b–c and Supplementary Figure 2c–d. b–c, Fluorescence emission spectra showing ligand-induced conformational changes of monobromobimane labeled β2AR reconstituted into high density lipoprotein particles (mBB-β2AR/HDL) in the absence (black solid line) or presence of full agonist isoproterenol (ISO, green wide dashed line), inverse agonist ICI-118,551 (ICI, black dashed line), Gs heterotrimer (red solid line), nanobody-80 (Nb80, blue solid lines), and combinations of Gs with ISO (red wide dashed line), Nb80 with ISO (blue wide dashed line), and Nb80 with ICI (blue dashed line). d–f Ligand binding curves for ISO competing against [3H]-dihydroalprenolol ([3H]-DHA) for d, β2AR/HDL reconstituted with Gs heterotrimer in the absence or presence GTPγS, e, β2AR/HDL in the absence and presence of Nb80, and f, β2AR-T4L/HDL in the absence and presence of Nb80. Error bars represent standard errors.
Figure 2. Comparison of the agonist-Nb80 stabilized crystal structures of the β2AR with inverse agonist bound β2AR and opsin
The structure of inverse agonist carazolol bound β2AR-T4L (β2AR-Cz) is shown in blue with the carazolol in yellow. The structure of BI-167107 agonist bound and Nb80 stabilized β2AR-T4L (β2AR-Nb80) is shown in orange with BI-167107 in green. These two structures were aligned using Pymol align function. a, Side view of the β2AR-Nb80 complex with β2AR in orange and CDRs of Nb80 in light blue (CDR1) and blue (CDR3). b, Side view of the superimposed structures showing significant structural changes in the intracellular and G protein facing part of the receptors. c, Comparison of the extracellular ligand binding domains showing modest structural changes. d, Cytoplasmic view showing the ionic lock interaction between Asp3.49 and Arg3.50 of the DRY motif in TM3 is broken in the β2AR-Nb80 structure. The intracellular end of TM6 is moved outward and away from the core of the receptor. The arrow indicates a 11.4 Å change in distance between the α-carbon of Glu6.30 in the structures of β2AR-Cz and β2AR-Nb80. The intracellular ends of TM3 and TM7 move towards the core by 4 and 2.5 Å respectively, while TM5 moves outward by 6Å. e, The β2AR-Nb80 structure superimposed with the structure of opsin crystallized with the C-terminal peptide of Gt (transducin) . PyMOL (
) was used for the preparation of all structure figures.
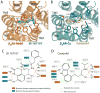
Figure 3. Ligand binding pocket of BI-167107 and carazolol bound β2AR structures
Panels a and b depict extracellular views of the agonist BI-167107 and carazolol bound structures, respectively. Residues within 4Å of one or both ligands are shown as sticks. In all panels, oxygens are red and nitrogens are blue. Panels c and d show a schematic representation of the interactions between the β2AR and the ligands BI-167107 and carazolol. The residues shown here have at least one atom within 4 Å of the ligand in the crystal structures. Mutations of amino acids in orange boxes have been shown to disrupt both antagonist and agonist binding. Mutations of amino acids in blue boxes have been shown to disrupt agonist binding. Green lines indicate potential hydrophobic interactions and orange lines indicate potential polar interactions.
Figure 4. Rearrangement of transmembrane segment packing interactions upon agonist binding
a, The BI-167107 and carazolol bound structures are superimposed to show structural differences propagating from the ligand binding pocket. BI-167107 and carazolol are shown with green and yellow bonds, respectively. b, Packing interactions that stabilize the inactive state are observed between Pro211 in TM5, Ile121 in TM3, Phe282 in TM6 and Asn318 in TM7. c, The inward movement of TM5 upon agonist binding disrupts the packing of Ile121 and Pro211 resulting in a rearrangement of interactions between Ile121 and Phe282. These changes contribute to a rotation and outward movement of TM6 and an inward movement of TM7.
Comment in
- Cell signalling: Binding the receptor at both ends.
Sprang SR. Sprang SR. Nature. 2011 Jan 13;469(7329):172-3. doi: 10.1038/469172a. Nature. 2011. PMID: 21228868 Free PMC article. - G protein-coupled receptors: Crystallizing how agonists bind.
Tse MT. Tse MT. Nat Rev Drug Discov. 2011 Feb;10(2):97. doi: 10.1038/nrd3379. Nat Rev Drug Discov. 2011. PMID: 21283100 No abstract available.
Similar articles
- Structure and function of an irreversible agonist-β(2) adrenoceptor complex.
Rosenbaum DM, Zhang C, Lyons JA, Holl R, Aragao D, Arlow DH, Rasmussen SG, Choi HJ, Devree BT, Sunahara RK, Chae PS, Gellman SH, Dror RO, Shaw DE, Weis WI, Caffrey M, Gmeiner P, Kobilka BK. Rosenbaum DM, et al. Nature. 2011 Jan 13;469(7329):236-40. doi: 10.1038/nature09665. Nature. 2011. PMID: 21228876 Free PMC article. - Molecular dynamics simulations of the effect of the G-protein and diffusible ligands on the β2-adrenergic receptor.
Goetz A, Lanig H, Gmeiner P, Clark T. Goetz A, et al. J Mol Biol. 2011 Dec 9;414(4):611-23. doi: 10.1016/j.jmb.2011.10.015. Epub 2011 Oct 20. J Mol Biol. 2011. PMID: 22037586 - Adrenaline-activated structure of β2-adrenoceptor stabilized by an engineered nanobody.
Ring AM, Manglik A, Kruse AC, Enos MD, Weis WI, Garcia KC, Kobilka BK. Ring AM, et al. Nature. 2013 Oct 24;502(7472):575-579. doi: 10.1038/nature12572. Epub 2013 Sep 22. Nature. 2013. PMID: 24056936 Free PMC article. - Structural features of β2 adrenergic receptor: crystal structures and beyond.
Bang I, Choi HJ. Bang I, et al. Mol Cells. 2015;38(2):105-11. doi: 10.14348/molcells.2015.2301. Epub 2014 Dec 24. Mol Cells. 2015. PMID: 25537861 Free PMC article. Review. - Agonist-bound structures of G protein-coupled receptors.
Lebon G, Warne T, Tate CG. Lebon G, et al. Curr Opin Struct Biol. 2012 Aug;22(4):482-90. doi: 10.1016/j.sbi.2012.03.007. Epub 2012 Apr 3. Curr Opin Struct Biol. 2012. PMID: 22480933 Review.
Cited by
- Advances in receptor conformation research: the quest for functionally selective conformations focusing on the β2-adrenoceptor.
Woo AY, Song Y, Zhu W, Xiao RP. Woo AY, et al. Br J Pharmacol. 2015 Dec;172(23):5477-88. doi: 10.1111/bph.13049. Epub 2015 Feb 27. Br J Pharmacol. 2015. PMID: 25537131 Free PMC article. Review. - Molecular recognition of ketamine by a subset of olfactory G protein-coupled receptors.
Ho J, Perez-Aguilar JM, Gao L, Saven JG, Matsunami H, Eckenhoff RG. Ho J, et al. Sci Signal. 2015 Mar 31;8(370):ra33. doi: 10.1126/scisignal.2005912. Sci Signal. 2015. PMID: 25829447 Free PMC article. - Ring current shifts in (19)F-NMR of membrane proteins.
Liu D, Wüthrich K. Liu D, et al. J Biomol NMR. 2016 May;65(1):1-5. doi: 10.1007/s10858-016-0022-4. Epub 2016 May 30. J Biomol NMR. 2016. PMID: 27240587 - The emerging mutational landscape of G proteins and G-protein-coupled receptors in cancer.
O'Hayre M, Vázquez-Prado J, Kufareva I, Stawiski EW, Handel TM, Seshagiri S, Gutkind JS. O'Hayre M, et al. Nat Rev Cancer. 2013 Jun;13(6):412-24. doi: 10.1038/nrc3521. Epub 2013 May 3. Nat Rev Cancer. 2013. PMID: 23640210 Free PMC article. Review. - Harvesting and cryo-cooling crystals of membrane proteins grown in lipidic mesophases for structure determination by macromolecular crystallography.
Li D, Boland C, Aragao D, Walsh K, Caffrey M. Li D, et al. J Vis Exp. 2012 Sep 2;(67):e4001. doi: 10.3791/4001. J Vis Exp. 2012. PMID: 22971942 Free PMC article.
References
- Scheerer P, et al. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. - PubMed
- Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454:183–U133. - PubMed
- Li J, Edwards PC, Burghammer M, Villa C, Schertler GF. Structure of bovine rhodopsin in a trigonal crystal form. J Mol Biol. 2004;343:1409–1438. - PubMed
- Palczewski K, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P60DK-20572/DK/NIDDK NIH HHS/United States
- GM083118/GM/NIGMS NIH HHS/United States
- R37 NS028471-21/NS/NINDS NIH HHS/United States
- R01 GM083118/GM/NIGMS NIH HHS/United States
- R01 GM083118-04/GM/NIGMS NIH HHS/United States
- R01 NS028471/NS/NINDS NIH HHS/United States
- NS028471/NS/NINDS NIH HHS/United States
- P01 GM75913/GM/NIGMS NIH HHS/United States
- R01 GM056169/GM/NIGMS NIH HHS/United States
- R37 NS028471/NS/NINDS NIH HHS/United States
- GM56169/GM/NIGMS NIH HHS/United States
- R01 GM068603/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials