The serine/threonine kinase Par1b regulates epithelial lumen polarity via IRSp53-mediated cell-ECM signaling - PubMed (original) (raw)
The serine/threonine kinase Par1b regulates epithelial lumen polarity via IRSp53-mediated cell-ECM signaling
David Cohen et al. J Cell Biol. 2011.
Abstract
The serine/threonine kinase Par1b promotes cell-cell adhesion and determines the polarity of the luminal domain in epithelial cells. In this study, we demonstrate that Par1b also regulates cell-extracellular matrix (ECM) signaling in kidney-derived Madin-Darby canine kidney (MDCK) cells and identified the rho-guanosine triphosphatase adaptor and scaffolding protein IRSp53 as a Par1b substrate involved in this pathway. Par1b overexpression inhibits basal lamina formation, cell spreading, focal adhesion, stress fiber formation, and compaction, whereas Par1b depletion has the opposite effect. IRSp53 depletion mimics Par1b overexpression on cell-ECM signaling and lumen polarity but had no effect on adherens junction formation. Par1b directly phosphorylates IRSp53 on S366 in cell lysates and stimulates phosphorylation on S453/3/5 via an indirect mechanism. A Par1b phosphorylation-deficient IRSp53 mutant but not the wild-type protein efficiently rescues both the cell spreading and the lumen polarity defects in Par1b MDCK cells. Our data suggest a model in which Par1b phosphorylation prevents recruitment of IRSp53 effector proteins to its Src homology domain 3 by promoting 14-3-3 binding in the vicinity of that domain.
Figures
Figure 1.
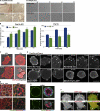
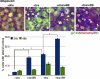
Par1b inhibits cell spreading and stress fiber formation and promotes cell compaction. (A) Phase-contrast images of cell islands cultured on tissue culture plastic for 24 h (left) or brightfield images of a time lapse (
Video 6
) after cell plating on collagen I (right). Translucent holes in the 24-h Par1b (−dox) culture representing lateral lumina. (B) Spreading on collagen I. *, P < 0.001 for differences between ±dox. Error bars indicate SEM. (C, D, and F) Still images from time-lapse videos 30 (C and F) or 45 min (D) after plating on collagen. (C) Brightfield and mRFP fluorescence (
Videos 1
and
2
) are shown. Arrows point to stress fibers. (D) Note the dox-dependent size difference in Par1b cells and Paxillin-mCherry (
Video 3
). Arrowheads point to FAs. (F) GFP or Par1b-GFP and mRFP actin (
Video 5
) are shown. Note both the difference in stress fibers (arrows) and that Par1b-GFP but not GFP localizes to the tips of actin bundles in lamella (arrowheads). (E) Polarized Par1b cultures 24 h after Ca2+ switch. Phalloidin labels stress fibers at the basal domain (bottom) and microvilli-rich lateral lumina and apical domains in the maximum (max) projections (top; Video 6). (G) Confocal sections through collagen cysts of Par1b-MDCK cells (±dox; see
Video 4
for full stacks). Arrowheads point to gaps in laminin. Also note that Par1b cysts are smaller than WT cysts (mean: 23 vs. 30 cells/cyst; n = 13/17). Bars: (A–F) 10 µm; (G) 25 µm.
Figure 2.
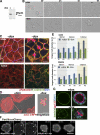
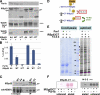
Par1b promotes IRSp53 phosphorylation and 14-3-3 binding. (A) IRSp53 in total lysates from Par1b and Par1b-KD cells (1/10 input) and from lysate fractions adsorbed on immobilized 14-3-3τ and competitively eluted. Arrows show a decrease in IRSp53 mobility in the Par1b lysates. (B) In vitro kinase assays of recombinant IRSp53 with Par1b-WT or Par1b-KA followed by λ-phosphatase (+) or mock (m) treatment and subsequent binding to immobilized 14-3-3τ. (top) 1/10 of IRSp53 input is shown. (middle) IB is shown, with IRSp53 competitively eluted from 14-3-3 resin. (bottom) Autorad is shown, with [32P]γATP-Par1b–phosphorylated IRSp53. (C) IRSp53-HA or IRSp53ΔSH3-HA cells transduced with control, Par1b-WT, or Par1b-KA adenovirus. IB of 1/10 Par1b, IB of HA input, and phospho-serine after HA IP are shown. (D) IRSp53-HA immunoprecipitated from [32P]orthophosphate-labeled (autorad) and unlabeled (IB, IRSp53-HA) Par1b-KD cells. Par1b IB shows dox-dependent depletion of the two Par1b splice variants.
Figure 3.
Par1 phosphorylation on S366 and Par1b-depedendent S453/4/5 phosphorylation promote 14-3-3 binding of IRSp53. (A) Diagram of Par1b-dependent phosphorylation sites in IRSp53. (B) In vitro kinase assays with WT and Par1b-KA using [32P]γATP and IRSp53-CT (WT and point mutants) as substrates. (C) IRSp53-HA–transfected cells (WT, A4, and S453-5A) were transduced with control (c) or Par1b virus, and the HA IPs were probed for P-serine and subsequently for HA. (D) In vitro kinase assays with WT and Par1b-ATP* using [35S]benzyl-ATPγS and full-length IRSp53 WT and mutants as substrates. Autoradiographs/Coomassie stain of IRSp53. (E) Triton X-100 lysates of cells coexpressing Par1b-WT or Par1b-ATP* and WT IRSp53-Myc or IRSp53 mutants were incubated with [35S]benzyl-ATPγS. Autoradiographs and IB for IRSp53-Myc and Par1b. (F) Recombinant WT IRSp53 and mutant proteins subjected to in vitro kinase assays with Par1b-WT or Par1b-KA were affinity isolated on 14-3-3 beads and competitively eluted (top). IRSp53 input (middle) and Par1b levels (bottom) are also shown. (G) 14-3-3 binding of IRSp53 proteins from cell lysates. Note the lower expression of the S117A sample accounts for the weaker signal in the bound fraction. The graph shows SEM from six experiments (three experiments for S117A/S148A). (H) Co-IP of 14-3-3-HA with IRSp53-Myc proteins from cell lysates.
Figure 4.
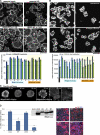
IRSp53 depletion mimics aspects of the Par1b overexpression phenotype. (A) Dox-inducible IRSp53 depletion is shown. (B) Brightfield time lapse of IRSp53-KD cell islands on collagen I (
Video 10
). (C) Polarized cultures 24 h after Ca2+ switch. Phalloidin labels stress fibers at the basal domain (bottom) and microvilli-rich lateral lumina and apical domains in the maximum (max) projections (top; see
Video 7
). (D and F) Still images from time-lapse videos taken 30 min after plating on collagen. (D) Brightfield and mRFP fluorescence (
Video 8
). (F) Paxillin-mCherry (
Video 9
, top) is shown. Arrowheads point to FAs. (E) Spreading of two IRSp53-KD clones on collagen I in DME and SMEM; all differences between ±dox are significant with P < 0.001. Error bars indicate SEM. (G) Confocal section through collagen cysts (see
Video 4
for full stacks) annotated as in Fig. 1 G. IRSp53KD cysts are smaller than control cysts (14 vs. 25 cells/cyst; n = 31). Bars: (B–D and F) 10 µm; (G) 25 µm.
Figure 5.
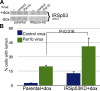
A nonphosphorylatable IRSp53 mutant rescues the Par1b phenotype. (A) Parental MDCK cells and five clones expressing RNAi-resistant IRSp53-A4 and four clones of IRSp53-WT cells were transiently transfected with mock or IRSp53shmir cDNA and analyzed for cell spreading 2 h after plating. (B) 2-h spreading in parental cells, eight IRSp53AA, and five IRSp53WT clones that were transduced with control or Par1b adenovirus. (C) Paxillin-mCherry in WT clone #1 and A4 clone #5 expressing Par1b-GFP 30 min after plating on collagen I. Arrowheads point to FAs (
Video 9
, bottom). (D) Par1b-MDCK cells transfected with mock or IRSp53 cDNAs (WT, A4, DD, and ΔSH3) were quantitatively analyzed for lumen polarity as described in Materials and methods. *, P < 0.0001 with difference to parental or mock-transfected cells. Error bars indicate SEM. Bars: (A and B) 30 µm; (C and D) 10 µm.
Figure 6.
Decreased IRSp53 and increased Par1b levels have a synergistic effect on lateral lumen formation. (A) IRSp53 expression in parental cells and an IRSp53KD clone in ±dox and after adenovirus transduction as indicated. (B) Lateral lumen polarity analyzed as described in Materials and methods. Error bars indicate SEM.
Figure 7.
IRSp53 and myosin II function synergistically to regulate epithelial lumen polarity. IRSp53KD (−dox) and control (+dox) subjected to Ca2+ switch assays for 24 and 48 h in the presence or absence of 50 µM BB. Lumen polarity was quantified as described in Materials and methods. Statistically significant difference is shown with P < 0.0001 (*). Images represent the 24-h time point. Error bars indicate SEM. Bars, 10 µm.
Figure 8.
Par1b phosphorylation inhibits the recruitment of WAVE2 and other IRSp53-binding proteins to its C-terminal, SH3-containing domain. (A and B) WAVE2 co-IPs with IRSp53 in 293 cells expressing WAVE2-GFP, Myc-tagged IRSp53 constructs (WT, A4, or IRSp53 Δ317–521; ΔCT), and Par1b where indicated. 1/10 of the lysate was analyzed for WAVE and Par1b expression. (B) Data are from three experiments with and without recombinant Par1b and three experiments without recombinant Par1b only. Error bars indicate SEM. (C) Cdc42Q61L co-IPs with IRSp53-Myc constructs from 293 cells. Arrows indicate Cdc42, and asterisks indicate IgG H and L chains. Data are representative of three experiments. (D–F) GST-IRSp53CT was subjected to Par1b phosphorylation, immobilized on glutathione and incubated with 14-3-3τ as indicated. (D) Experimental schematic is shown. (E) Binding of proteins from [35S]Met/Cys-labeled 293 lysates to immobilized IRSp53CT was analyzed by Coomassie and autoradiography. Arrows indicate specific bands that are reduced in the Par1b sample. Asterisk indicates cellular 14-3-3, with one out of two identical results represented. (F) 14-3-3 binding to IRSp53CT was analyzed by IB, and IRSp53-CT levels by Ponceau red stain are shown.
Figure 9.
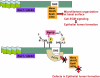
Model for Par1b regulation of IRSp53 in epithelial lumen formation. Activated Rac1 or Cdc42 recruit Rho-GTPase effector proteins such as Mena, WAVE, mDia, and Eps8 involved in microfilament organization to the IRSp53 SH3 domain. Phosphorylation of S366 by Par1b and Par1b stimulation of S434-5 phosphorylation by an unidentified kinase recruits 14-3-3 proteins that interfere with WAVE2 binding and the recruitment of other proteins that interact with the IRSp53 C-terminal domain. Par1b overexpression inhibits IRSp53-dependent actin dynamics that regulate cell–ECM signaling and result in altered lumen polarity in MDCK cells. In other cell types, such as Cos7, unidentified kinases inhibit IRSp53 function by creating 14-3-3 binding sites on T340/T360.
Similar articles
- Par1b links lumen polarity with LGN-NuMA positioning for distinct epithelial cell division phenotypes.
Lázaro-Diéguez F, Cohen D, Fernandez D, Hodgson L, van Ijzendoorn SC, Müsch A. Lázaro-Diéguez F, et al. J Cell Biol. 2013 Oct 28;203(2):251-64. doi: 10.1083/jcb.201303013. J Cell Biol. 2013. PMID: 24165937 Free PMC article. - Par1b promotes hepatic-type lumen polarity in Madin Darby canine kidney cells via myosin II- and E-cadherin-dependent signaling.
Cohen D, Tian Y, Müsch A. Cohen D, et al. Mol Biol Cell. 2007 Jun;18(6):2203-15. doi: 10.1091/mbc.e07-02-0095. Epub 2007 Apr 4. Mol Biol Cell. 2007. PMID: 17409351 Free PMC article. - PAR1b promotes cell-cell adhesion and inhibits dishevelled-mediated transformation of Madin-Darby canine kidney cells.
Elbert M, Cohen D, Müsch A. Elbert M, et al. Mol Biol Cell. 2006 Aug;17(8):3345-55. doi: 10.1091/mbc.e06-03-0193. Epub 2006 May 17. Mol Biol Cell. 2006. PMID: 16707567 Free PMC article. - Apical surface formation in MDCK cells: regulation by the serine/threonine kinase EMK1.
Cohen D, Müsch A. Cohen D, et al. Methods. 2003 Jul;30(3):269-76. doi: 10.1016/s1046-2023(03)00033-1. Methods. 2003. PMID: 12798141 Review. - Molecular regulation of lumen morphogenesis.
Datta A, Bryant DM, Mostov KE. Datta A, et al. Curr Biol. 2011 Feb 8;21(3):R126-36. doi: 10.1016/j.cub.2010.12.003. Curr Biol. 2011. PMID: 21300279 Free PMC article. Review.
Cited by
- Hepatocyte polarity.
Treyer A, Müsch A. Treyer A, et al. Compr Physiol. 2013 Jan;3(1):243-87. doi: 10.1002/cphy.c120009. Compr Physiol. 2013. PMID: 23720287 Free PMC article. Review. - Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Fedoryshchak RO, Přechová M, Butler AM, Lee R, O'Reilly N, Flynn HR, Snijders AP, Eder N, Ultanir S, Mouilleron S, Treisman R. Fedoryshchak RO, et al. Elife. 2020 Sep 25;9:e61509. doi: 10.7554/eLife.61509. Elife. 2020. PMID: 32975518 Free PMC article. - Impact of HCV Infection on Hepatocyte Polarity and Plasticity.
Agnetti J, Desterke C, Gassama-Diagne A. Agnetti J, et al. Pathogens. 2022 Mar 10;11(3):337. doi: 10.3390/pathogens11030337. Pathogens. 2022. PMID: 35335661 Free PMC article. Review. - The signaling adaptor GAB1 regulates cell polarity by acting as a PAR protein scaffold.
Yang Z, Xue B, Umitsu M, Ikura M, Muthuswamy SK, Neel BG. Yang Z, et al. Mol Cell. 2012 Aug 10;47(3):469-83. doi: 10.1016/j.molcel.2012.06.037. Mol Cell. 2012. PMID: 22883624 Free PMC article. - Par1b links lumen polarity with LGN-NuMA positioning for distinct epithelial cell division phenotypes.
Lázaro-Diéguez F, Cohen D, Fernandez D, Hodgson L, van Ijzendoorn SC, Müsch A. Lázaro-Diéguez F, et al. J Cell Biol. 2013 Oct 28;203(2):251-64. doi: 10.1083/jcb.201303013. J Cell Biol. 2013. PMID: 24165937 Free PMC article.
References
- Blethrow J., Zhang C., Shokat K., Weiss E. 2004. Design and use of analog-sensitive protein kinases. Curr. Protoc. Mol. Biol. Unit 18.11 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous