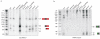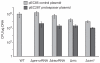CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III - PubMed (original) (raw)
CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III
Elitza Deltcheva et al. Nature. 2011.
Abstract
CRISPR/Cas systems constitute a widespread class of immunity systems that protect bacteria and archaea against phages and plasmids, and commonly use repeat/spacer-derived short crRNAs to silence foreign nucleic acids in a sequence-specific manner. Although the maturation of crRNAs represents a key event in CRISPR activation, the responsible endoribonucleases (CasE, Cas6, Csy4) are missing in many CRISPR/Cas subtypes. Here, differential RNA sequencing of the human pathogen Streptococcus pyogenes uncovered tracrRNA, a trans-encoded small RNA with 24-nucleotide complementarity to the repeat regions of crRNA precursor transcripts. We show that tracrRNA directs the maturation of crRNAs by the activities of the widely conserved endogenous RNase III and the CRISPR-associated Csn1 protein; all these components are essential to protect S. pyogenes against prophage-derived DNA. Our study reveals a novel pathway of small guide RNA maturation and the first example of a host factor (RNase III) required for bacterial RNA-mediated immunity against invaders.
Figures
Figure 1. A newly identified tracrRNA is required for crRNA maturation in S. pyogenes
a, dRNA-seq reveals expression of tracrRNA and crRNAs. Sequence reads of cDNA libraries derived from untreated and TEX-treated total RNA are shown. Vertical axis, relative amounts of sequenced cDNAs. The absence of ~75 nt tracrRNA form and 39-42 nt crRNA fragments in the TEX-treated cDNA library indicates that they are generated by processing. Genomic organization of tracrRNA and CRISPR01/Cas (csn1-cas1-cas2-csn2) loci. Transcriptional start sites and a terminator are indicated. (Left) tracrRNA (red) is encoded on the minus strand and detected as 171, 89 and ~75 nt tracrRNA species. Black rectangle, 36 nt sequence stretch complementary to CRISPR01 repeat. (Right) pre-crRNA is encoded on the plus strand. Rectangles, CRISPR01 repeats; diamonds, CRISPR01 spacers; 511, 66 and 39-42 nt, pre-crRNA and processed crRNAs. b, Base-pairing of tracrRNA with a CRISPR01 repeat is represented. Cleavages observed by dRNA-seq and leading to the formation of short overhangs at the 3′ ends of the processed RNAs are indicated by black arrows. Open arrow, cleavage in the spacer sequence. c, tracrRNA and pre-crRNA are co-processed in vivo. Northern blot analysis of S. pyogenes total RNA: strains and probes are indicated (Supplementary Figs 2 and 4). Left panel: Processing of tracrRNA into the ~75 nt form is abolished in Δpre-crRNA and re-established upon complementation with pre-crRNA. Right panel: Processing of pre-crRNA into mature crRNA forms (39-42 nt) is abrogated in ΔtracrRNA. Trans complementation of ΔtracrRNA with 171 or 89 nt tracrRNA restores the processing.
Figure 2. Co-processing of tracrRNA and pre-crRNA requires both endogenous RNase III and Csn1 in vivo
Northern blot analysis of tracrRNA (a) and pre-crRNA (b) expression: strains and probes are indicated (Supplementary Figs 6 and 8). Processing of tracrRNA (a) into a ~75 nt form and pre-crRNA (b) into 39-42 nt mature crRNA forms is abolished in Δ_rnc_, Δ_cas/csn_ and Δ_csn1_ (refer to Supplementary Figs 6 and 8).
Figure 3. tracrRNA directs pre-crRNA cleavage by RNase III in vitro
a, Schematic representation of tracrRNA89 corresponding to 89-nt long tracrRNA, and crRNA213 and crRNA148 corresponding to a 213-nt long leader-repeat-spacer1-repeat-spacer2 fragment and a 148-nt long spacer1-repeat-spacer2-repeat-spacer3 fragment, respectively. b, Identification of tracrRNA89 binding sites on crRNA148*. 5′ end-labeled crRNA148* (~10 nM) was subjected to lead(II), RNase III and RNase T1 cleavage in the absence (lanes 1, 4, 7) and presence of cold tracrRNA89 (final concentration in lanes 2, 5, and 7: ~50 nM; lanes 3, 6, and 9: ~500 nM). Lane C: untreated crRNA148*; Lane T1: RNase T1 digest of crRNA148* under denaturating conditions; Lane OH: alkaline ladder; cleaved G residues are labeled. Vertical bars: crRNA148 region protected by tracrRNA89. Arrows denote specific RNase III cleavages in the two repeat regions of crRNA148 in the presence of tracrRNA89. c, Identification of crRNA148 and crRNA213 binding sites on tracrRNA89. 5′ end-labeled tracrRNA89* (~10 nM) was subjected to RNase T1, lead(II) and RNase III cleavage in the absence (lanes 1, 6, 11) and presence of cold crRNA148 or crRNA213 (final concentration in lanes 2, 4, 7, 9, 12 and 14: ~50 nM; lanes 3, 5, 8, 10, 13 and 15: ~500 nM). Lanes C, T1 and OH, positions of cleaved Gs and vertical bars: as above but referring to tracrRNA89* in the presence of cold crRNA148 or crRNA213.
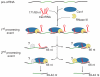
Figure 4. Model for tracrRNA-mediated crRNA maturation involving RNase III and Csn1
Black, repeat; green, spacer. tracrRNA can bind with almost perfect complementarity to each repeat sequence of the pre-crRNA. The resulting RNA duplex is recognized and site-specifically diced by RNase III in the presence of Csn1, releasing the individual repeat-spacer-repeat units (1st processing event). The generated units undergo further processing within the spacer sequences resulting in mature crRNA species consisting of unique spacer-repeat sequences (2nd processing event) by a yet-to-be elucidated mechanism. Csn1 may also be involved in the silencing of invading sequences.
Figure 5. Both tracrRNA and pre-crRNA confer immunity against acquisition of a protospacer gene derived from a lysogenic phage
Transformation efficiencies of S. pyogenes with speM protospacer containing plasmid (pEC287) and reference “backbone” plasmid (pEC85) (Supplementary Fig. 9). Strains: S. pyogenes WT (SF370), Δpre-crRNA, ΔtracrRNA, Δ_rnc_ and Δ_csn1_. Graph bars, mean values of colony forming units (CFU) per μg of plasmid DNA; error bars, standard deviation (SD); n≥3. pEC287 is tolerated by the Δpre-crRNA, ΔtracrRNA, Δ_rnc_ and Δ_csn1_ mutants but not by the WT strain. As control, transformants in all strains were obtained with the backbone plasmid (Supplementary Fig. 9).
Figure 6. tracrRNA-mediated crRNA maturation is conserved among different bacterial species
tracrRNA-mediated crRNA maturation is inherent to the type II (Nmeni/CASS4) CRISPR/Cas systems. Type II (Nmeni/CASS4) loci from S. pyogenes SF370, S. mutans UA159, L. innocua Clip11262, N. meningitidis Z2491 and S. thermophilus LMD-9 (Nmeni/CASS4a); red, tracrRNA; rectangles, repeats; diamonds, spacers.
Comment in
- Microbiology: Dicing defence in bacteria.
Gottesman S. Gottesman S. Nature. 2011 Mar 31;471(7340):588-9. doi: 10.1038/471588a. Nature. 2011. PMID: 21455171 No abstract available.
Similar articles
- Microbiology: Dicing defence in bacteria.
Gottesman S. Gottesman S. Nature. 2011 Mar 31;471(7340):588-9. doi: 10.1038/471588a. Nature. 2011. PMID: 21455171 No abstract available. - The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems.
Chylinski K, Le Rhun A, Charpentier E. Chylinski K, et al. RNA Biol. 2013 May;10(5):726-37. doi: 10.4161/rna.24321. Epub 2013 Apr 5. RNA Biol. 2013. PMID: 23563642 Free PMC article. - The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA.
Fonfara I, Richter H, Bratovič M, Le Rhun A, Charpentier E. Fonfara I, et al. Nature. 2016 Apr 28;532(7600):517-21. doi: 10.1038/nature17945. Epub 2016 Apr 20. Nature. 2016. PMID: 27096362 - Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR-Cas adaptive immunity.
Charpentier E, Richter H, van der Oost J, White MF. Charpentier E, et al. FEMS Microbiol Rev. 2015 May;39(3):428-41. doi: 10.1093/femsre/fuv023. Epub 2015 May 19. FEMS Microbiol Rev. 2015. PMID: 25994611 Free PMC article. Review. - The tracrRNA in CRISPR Biology and Technologies.
Liao C, Beisel CL. Liao C, et al. Annu Rev Genet. 2021 Nov 23;55:161-181. doi: 10.1146/annurev-genet-071719-022559. Epub 2021 Aug 20. Annu Rev Genet. 2021. PMID: 34416117 Free PMC article. Review.
Cited by
- Choosing the Right Tool for the Job: RNAi, TALEN, or CRISPR.
Boettcher M, McManus MT. Boettcher M, et al. Mol Cell. 2015 May 21;58(4):575-85. doi: 10.1016/j.molcel.2015.04.028. Mol Cell. 2015. PMID: 26000843 Free PMC article. Review. - Survival and Evolution of CRISPR-Cas System in Prokaryotes and Its Applications.
Shabbir MA, Hao H, Shabbir MZ, Hussain HI, Iqbal Z, Ahmed S, Sattar A, Iqbal M, Li J, Yuan Z. Shabbir MA, et al. Front Immunol. 2016 Sep 26;7:375. doi: 10.3389/fimmu.2016.00375. eCollection 2016. Front Immunol. 2016. PMID: 27725818 Free PMC article. Review. - High-temperature protein G is essential for activity of the Escherichia coli clustered regularly interspaced short palindromic repeats (CRISPR)/Cas system.
Yosef I, Goren MG, Kiro R, Edgar R, Qimron U. Yosef I, et al. Proc Natl Acad Sci U S A. 2011 Dec 13;108(50):20136-41. doi: 10.1073/pnas.1113519108. Epub 2011 Nov 23. Proc Natl Acad Sci U S A. 2011. PMID: 22114197 Free PMC article. - Development of an Efficient Genome Editing Method by CRISPR/Cas9 in a Fish Cell Line.
Dehler CE, Boudinot P, Martin SA, Collet B. Dehler CE, et al. Mar Biotechnol (NY). 2016 Aug;18(4):449-52. doi: 10.1007/s10126-016-9708-6. Epub 2016 May 28. Mar Biotechnol (NY). 2016. PMID: 27236514 Free PMC article. - Editing of the Bacillus subtilis Genome by the CRISPR-Cas9 System.
Altenbuchner J. Altenbuchner J. Appl Environ Microbiol. 2016 Aug 15;82(17):5421-7. doi: 10.1128/AEM.01453-16. Print 2016 Sep 1. Appl Environ Microbiol. 2016. PMID: 27342565 Free PMC article.
References
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: a Laboratory Manual. 2nd edn. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N. Y.: 1989. edn.
- Caparon MG, Scott JR. Genetic manipulation of pathogenic streptococci. Methods Enzymol. 1991;204:556–586. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous