A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis - PubMed (original) (raw)
A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis
Gira Bhabha et al. Science. 2011.
Abstract
Conformational dynamics play a key role in enzyme catalysis. Although protein motions have clear implications for ligand flux, a role for dynamics in the chemical step of enzyme catalysis has not been clearly established. We generated a mutant of Escherichia coli dihydrofolate reductase that abrogates millisecond-time-scale fluctuations in the enzyme active site without perturbing its structural and electrostatic preorganization. This dynamic knockout severely impairs hydride transfer. Thus, we have found a link between conformational fluctuations on the millisecond time scale and the chemical step of an enzymatic reaction, with broad implications for our understanding of enzyme mechanisms and for design of novel protein catalysts.
Figures
Fig. 1
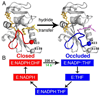
Conformational changes that occur during the E. coli DHFR catalytic cycle. (A) Left, cartoon representation of E:NADP+:FOL crystal structure (1RX2, model for the Michaelis complex, E:NADPH:DHF) in the closed conformation. Right, crystal structure of E:NADP+:ddTHF (1RX4, model for the product complex) in the occluded conformation. NADP+ is shown in orange; FOL is shown in yellow and ddTHF in magenta. Red, Met20 loop in the closed conformation; blue, Met20 loop in the occluded conformation. The sites of mutation, N23 and S148 are shown as spheres. (B) Intermediates in the wild type E. coli DHFR catalytic cycle. Intermediates shown in red are in the closed conformation, and those in blue are in the occluded conformation. Prior to hydride transfer the Met20 loop is in the closed conformation, where it packs tightly against the nicotinamide ring of NADP+. Following hydride transfer, the Met20 loop adopts the occluded conformation, in which the nicotinamide ring of NADP+ is sterically hindered from binding in the active site. NADP+ undergoes a concurrent conformational change in which the nicotinamide ring is expelled from the binding pocket, initiating NADP+ release from the ternary product complex. The rate of hydride transfer in the wild type and N23PP/S148A mutant enzyme is indicated in black and green, respectively. Note that the mutation alters the pathway utilized for product and NADP+ release, as shown in Fig. S2.
Fig. 2
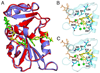
The three-dimensional structures of the E:NADP+:FOL complexes of the N23PP/S148A mutant and wild-type E. coli DHFR are almost identical. (A) Superposition of the crystal structures of wild-type E. coli DHFR (1RX2) and N23PP/S148A E. coli DHFR. Wild-type E. coli DHFR is shown in red, with yellow ligands and N23PP/S148A E. coli DHFR is shown in purple, with green ligands. (B) Active site configuration for wild-type ecDHFR (1RX2-re-refined). Folate is colored yellow, NADP+ orange, the waters are shown as green spheres, and key active-site residues are in blue sticks. (C) Active site configuration for N23PP/S148A ecDHFR. Colors are the same as for B. For clarity, only the major conformation of the glutamate moiety of folate is shown. The active site configurations are almost identical for wild type and N23PP/S148A ecDHFR, including placement of polar residues, key hydrophobic residues and waters, showing that the electrostatic nature of the active site is unchanged by the mutations.
Fig. 3
The Met20 loop of N23PP/S148A E. coli DHFR remains in the closed position across the chemical step. (A) 1H-15N HSQC spectra of wild-type E:NADP+:FOL (model Michaelis complex, black) in the closed conformation and E:NADP+:THF (product complex, red) in the occluded conformation. Large chemical shift differences are observed, particularly for residues in regions that undergo the closed-to-occluded structural change. Chemical shift changes from closed (black) to occluded (red) are indicated by arrows for several residues in the active site loops. (B) 1H-15N HSQC spectra of N23PP/S148A E:NADP+:FOL (model Michaelis complex, black) and E:NADP+:THF (product complex, red). The 1H-15N HSQC spectra for the complexes of the mutant enzyme are similar; the cross peaks corresponding to the active site loops do not shift, and appear in the position corresponding to the closed conformation for both complexes. A quantitative chemical shift analysis is presented in Fig. S8.
Fig. 4
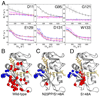
Millisecond timescale dynamics in the active site loops of the E:NADP+:FOL complex are impaired by the N23PP/S148A mutation. (A) Representative 15N R2 relaxation dispersion curves for wild type (black), N23PP/S148A (red), N23PP (blue) and S148A (magenta) E:NADP+:FOL. Dispersion data were collected at pH 7.6 and at 301 K. Top, active site loop residues (11, 95, 121), for which the dispersion observed in wild type E:NADP+:FOL is not observed in either N23PP/S148A or N23PP E:NADP+:FOL. S148A retains dispersion for residue 11 and other active site residues, shown in Fig. S11. Bottom, C-terminal associated residues (129, 131, 133), for which dispersion is observed for the wild type and mutant proteins. Residues which display 15N relaxation dispersion in the E:NADP+:FOL complex are mapped onto the structure for wild type (B), N23PP/S148A (C) and S148A (D). 1RX2 coordinates are used for representations in B and D. Red spheres indicate active site-associated residues for which R2 dispersion is observed. Blue spheres indicate C-terminal-associated residues for which R2 dispersion is observed. NADP+ is shown as orange sticks, FOL as yellow sticks. For clarity, only the major conformation of the glutamate moiety of folate is shown.
Similar articles
- The role of the Met20 loop in the hydride transfer in Escherichia coli dihydrofolate reductase.
Mhashal AR, Vardi-Kilshtain A, Kohen A, Major DT. Mhashal AR, et al. J Biol Chem. 2017 Aug 25;292(34):14229-14239. doi: 10.1074/jbc.M117.777136. Epub 2017 Jun 15. J Biol Chem. 2017. PMID: 28620051 Free PMC article. - Evidence that a 'dynamic knockout' in Escherichia coli dihydrofolate reductase does not affect the chemical step of catalysis.
Loveridge EJ, Behiry EM, Guo J, Allemann RK. Loveridge EJ, et al. Nat Chem. 2012 Mar 11;4(4):292-7. doi: 10.1038/nchem.1296. Nat Chem. 2012. PMID: 22437714 - Defining the role of active-site loop fluctuations in dihydrofolate reductase catalysis.
McElheny D, Schnell JR, Lansing JC, Dyson HJ, Wright PE. McElheny D, et al. Proc Natl Acad Sci U S A. 2005 Apr 5;102(14):5032-7. doi: 10.1073/pnas.0500699102. Epub 2005 Mar 28. Proc Natl Acad Sci U S A. 2005. PMID: 15795383 Free PMC article. - Distal Regions Regulate Dihydrofolate Reductase-Ligand Interactions.
Goldstein M, Goodey NM. Goldstein M, et al. Methods Mol Biol. 2021;2253:185-219. doi: 10.1007/978-1-0716-1154-8_12. Methods Mol Biol. 2021. PMID: 33315225 Review.
Cited by
- Integrative, dynamic structural biology at atomic resolution--it's about time.
van den Bedem H, Fraser JS. van den Bedem H, et al. Nat Methods. 2015 Apr;12(4):307-18. doi: 10.1038/nmeth.3324. Nat Methods. 2015. PMID: 25825836 Free PMC article. Review. - Heteronuclear Adiabatic Relaxation Dispersion (HARD) for quantitative analysis of conformational dynamics in proteins.
Traaseth NJ, Chao FA, Masterson LR, Mangia S, Garwood M, Michaeli S, Seelig B, Veglia G. Traaseth NJ, et al. J Magn Reson. 2012 Jun;219:75-82. doi: 10.1016/j.jmr.2012.03.024. Epub 2012 Apr 6. J Magn Reson. 2012. PMID: 22621977 Free PMC article. - Observing lysozyme's closing and opening motions by high-resolution single-molecule enzymology.
Akhterov MV, Choi Y, Olsen TJ, Sims PC, Iftikhar M, Gul OT, Corso BL, Weiss GA, Collins PG. Akhterov MV, et al. ACS Chem Biol. 2015 Jun 19;10(6):1495-501. doi: 10.1021/cb500750v. Epub 2015 Mar 20. ACS Chem Biol. 2015. PMID: 25763461 Free PMC article. - Revealing the peptide presenting process of human leukocyte antigen through the analysis of fluctuation.
Yanaka S, Ueno T, Tsumoto K, Sugase K. Yanaka S, et al. Biophysics (Nagoya-shi). 2015 Apr 18;11:103-6. doi: 10.2142/biophysics.11.103. eCollection 2015. Biophysics (Nagoya-shi). 2015. PMID: 27493522 Free PMC article. Review. - Photoconvertible Fluorescent Proteins and the Role of Dynamics in Protein Evolution.
Wachter RM. Wachter RM. Int J Mol Sci. 2017 Aug 18;18(8):1792. doi: 10.3390/ijms18081792. Int J Mol Sci. 2017. PMID: 32962314 Free PMC article.
References
- Hammes-Schiffer S, Benkovic SJ. Annu. Rev. Biochem. 2006;75:519. - PubMed
- Henzler-Wildman K, Kern D. Nature. 2007;450:964. - PubMed
- Wolf-Watz M, et al. Nat Struct Mol Biol. 2004;11:945. - PubMed
- Boehr DD, McElheny D, Dyson HJ, Wright PE. Science. 2006;313:1638. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- Y1-GM-1104/GM/NIGMS NIH HHS/United States
- GM75995/GM/NIGMS NIH HHS/United States
- R01 GM075995/GM/NIGMS NIH HHS/United States
- Y1-CO-1020/CO/NCI NIH HHS/United States
- GM080209/GM/NIGMS NIH HHS/United States
- U54 GM094586/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials