A Drosophila model of FUS-related neurodegeneration reveals genetic interaction between FUS and TDP-43 - PubMed (original) (raw)
. 2011 Jul 1;20(13):2510-23.
doi: 10.1093/hmg/ddr150. Epub 2011 Apr 12.
Affiliations
- PMID: 21487023
- PMCID: PMC4288133
- DOI: 10.1093/hmg/ddr150
A Drosophila model of FUS-related neurodegeneration reveals genetic interaction between FUS and TDP-43
Nicholas A Lanson Jr et al. Hum Mol Genet. 2011.
Abstract
Amyotrophic lateral sclerosis (ALS) is a late-onset neurodegenerative disorder characterized by the loss of motor neurons. Fused in sarcoma/translated in liposarcoma (FUS/TLS) and TAR DNA-binding protein (TDP)-43 are DNA/RNA-binding proteins found to be mutated in sporadic and familial forms of ALS. Ectopic expression of human ALS-causing FUS/TLS mutations in Drosophila caused an accumulation of ubiquitinated proteins, neurodegeneration, larval-crawling defect and early lethality. Mutant FUS/TLS localized to both the cytoplasm and nucleus, whereas wild-type FUS/TLS localized only to the nucleus, suggesting that the cytoplasmic localization of FUS/TLS is required for toxicity. Furthermore, we found that deletion of the nuclear export signal strongly suppressed toxicity, suggesting that cytoplasmic localization is necessary for neurodegeneration. Interestingly, we observed that FUS/TLS genetically interacts with TDP-43 in a mutation-dependent fashion to cause neurodegeneration in vivo. In summary, we demonstrate that ALS-associated mutations in FUS/TLS cause adult-onset neurodegeneration via a gain-of-toxicity mechanism that involves redistribution of the protein from the nucleus to the cytoplasm and is likely to involve an interaction with TDP-43.
Figures
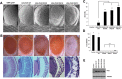
Figure 1.
Human ALS-causing mutations in FUS lead to neurodegeneration in Drosophila. (A) Scanning electron micrographs of Day 1 adult fly eyes in which expression of WT or mutant FUS is targeted by the eye-specific promoter GMR-GAL4. The eyes of control flies (GMR-GAL4/+) and flies expressing WT FUS are almost normal and have an organized ommatidial architecture [(A) and (B)]. The eyes of flies expressing mutant FUS R518K or R521H show ommatidial degeneration, partial collapse and loss of eye pigmentation. The eyes of flies expressing mutant FUS R521C are severely degenerated with complete loss of eye pigmentation. (B) Light micrographs of Day 1 adult fly eyes expressing WT or mutant FUS under the control of the GMR-GAL4 driver. Bottom row: corresponding Richardson's stained frontal sections showing a mutation-dependent severe internal degeneration. (C) Quantitative analysis of eye phenotypes. Flies from each genotype were randomly selected for objective scoring according to the criteria described previously (15). Flies carrying a mutated FUS R518K, R521H or R521C all display a more severe phenotype. Values are the means ± SEM. The severity of the phenotype between FUS-WT (*) and each of the individual mutants (**) was significant (P < 0.0001). (D) Ectopic expression of FUS in the fly neurons results in mutation-dependent loss of viability. WT or mutant (R518K, R521C and R521H) FUS was expressed by using the pan-neuronal driver Appl-GAL4. Mean eclosion rates were drastically reduced in pupae with pan-neuronal expression of mutant FUS R521H and no R518K or R521H flies eclosed. However, pupae expressing WT FUS eclosed normally when compared with the control (Appl-GAL4 driver alone). Values are the means ± SEM. Although control (*) and FUS-WT (*) are not significantly different from each other, they are each significantly different from each of the FUS mutants (**) (P < 0.0001). (E) Western blot showing expression levels of WT and mutant forms of FUS in the eyes of Day 1 adult flies (top panel). Tubulin served as a loading control (lower panel).
Figure 2.
Conditional expression of mutant FUS in the adult nervous system leads to a severe reduction in life span and climbing ability. Driver lines expressing the transcriptional activator Gene Switch under control of the neuron-specific promoter elav-GS were crossed with either UAS-FUS WT or UAS-FUS R521C flies. Following treatment with RU486, the Gene Switch protein is transcriptionally activated and binds to UAS and thus induces expression of the respective FUS protein specifically in the fly nervous system. (A and B) Lifespan of female and male flies expressing FUS WT and FUS R521C transgenes under the control of elav-GS. The survival curves show the percentage of living flies as a function of age. Neuronal expression of FUS R521C strongly reduces lifespan, whereas FUS WT expression has a significantly weaker effect on the reduction of life span. Control animals (Elav-GS/+) raised and maintained with RU486 (+) or without RU486 (−) showed normal life span with many flies surviving beyond the end of the study. (C) Expression of FUS R521C in the adult fly nervous system caused age-dependent and mutation-dependent locomotor dysfunction. Climbing ability of Elav-GS/+, UAS-FUS WT/+;elavGS/+ and UAS-R521C/+;elavGS/+ flies raised with RU486 (+) or without RU486 (−) were assessed at the indicated time-points. Neuronal expression of FUS R521C caused a severe locomotor defect when compared with age-matched FUS WT animals (<3% of the FUS R521C flies were able to climb 10 cm when compared with 41% of FUS WT expressing flies on Day 15). Control animals (Elav-GS/+) raised and maintained with RU486 (+) and without RU486 (−) showed normal climbing behavior. (D) Western blot analyses of total protein extracts prepared from heads of new born flies in which expression of UAS-FUS was induced (+) or not (−) with RU486 for 1 day and 15 days. As reported previously, a thin band of FUS is detected in the absence of RU486, indicating a leak in the control of expression (21). Expression of both WT and mutant FUS was significantly increased on Day 15. Tubulin was used as a loading control.
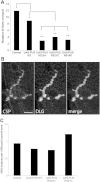
Figure 3.
Expression of mutant FUS in motor neurons leads to a larval crawling defect without affecting neuromuscular junction structure. (A) Ectopic expression of FUS R518K, FUS R521C and FUS R521H in motor neurons results in a larval crawling defect when compared with FUS WT expressing animals or driver alone. Values are the means ± SEM. FUS-WT (*) is significantly different from the control (*) (P < 0.0001). Both the control and FUS-WT are significantly different from each of the FUS mutants (**) (P < 0.0001). (B) Muscle 4, segment A3 larval neuromuscular junction of larva expressing FUS R521C under the control of OK371-GAL4 display normal morphology of synaptic boutons as determined by immunohistochemistry with antibodies against pre-synaptic (CSP) and post-synaptic (discs large, DLG) proteins. Scale bar is 10 µm. (C) FUS WT, FUS R521C and FUS R521H expressing animals have a normal number of synaptic boutons when compared with the non-transgenic animals. There is no significant difference between any of the groups. N = 15 synapses from four animals of each genotype.
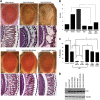
Figure 4.
FUS NES deletion (FUS ΔNES) rescues the degenerative phenotype associated with ALS-causing FUS mutations. (A) Light micrographs of eyes from 1-day-old flies in which FUS WT, FUS R518K and FUS R521C were expressed by using the GMR-GAL4 driver. Eyes from the line expressing FUS WT showed almost normal morphology by light microscopy and histology. However, the FUS R518K and FUS R521C lines showed moderate and severe eye degeneration, respectively. Ectopic expression of FUS ΔNES does not cause any external eye degeneration, similar to the FUS-WT line. Strikingly, deletion of the NES strongly suppressed neurodegeneration associated with R518K and R521C mutations. (B) Quantitative analysis of eye phenotypes. Flies from each genotype were randomly selected for objective scoring according to criteria described previously (15). While R518K and R521C induced severe degeneration of the eyes, the combination of these mutations with deletion of the NES resulted in an almost total reduction in severity. Values are the means ± SEM. The single asterisk denotes the significant difference between the indicated pairs (P < 0.001). The double asterisks denote the significant difference between the indicated pairs (P < 0.001). The difference between FUS WT and FUS ΔNES is less significant (P < 0.05). (C) Ectopic expression of FUS in fly neurons results in mutation-dependent loss of viability. WT or mutant (R518K, R521C or R521H) FUS was expressed by using the pan-neuronal driver Appl-GAL4. Deletion of the NES strongly suppressed the mutation-dependent viability defect and flies expressing FUS ΔNES R521C and FUS ΔNES R518K eclosed normally when compared with FUS WT and the driver alone. Mean eclosion rates were drastically reduced in pupae with pan-neuronal expression of either FUS R521C or R518K. Values are the means ± SEM. The single asterisk denotes the significant difference between the indicated pairs (P < 0.01 for each matching). There is no significant difference between FUS WT and FUS ΔNES (NS). (D) Western blot analysis of FUS expression in fly heads from FUS WT, FUS-R518K, FUS-R521C, FUS ΔNES, FUS ΔNES R518K and FUS ΔNES R521C expressing transgenic flies. Tubulin served as a loading control.
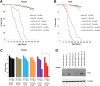
Figure 5.
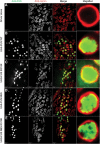
WT human FUS is predominantly nuclear, but mutant FUS is also redistributed to the cytoplasm. FUS mutants with deletion of the NES are retained exclusively in the nucleus. Lamin was used as a nuclear marker to highlight the nuclear envelope membrane, and thus immunostaining inside the lamin ring is nuclear and staining outside is cytoplasmic. (A) OK371-GAL4 (control) third instar larvae stained with both anti-FUS and anti-lamin antibody clearly showed a nuclear envelope membrane. The anti-FUS antibody resulted in a non-specific tiny punctuate staining pattern in control animals mostly outside the nucleus. (B) WT FUS protein was predominantly in the nucleus. (C) FUS R518K, besides the nucleus, was also redistributed to the cytoplasm. FUS R518K staining (C, arrows) was also in the cytoplasm, outside of the lamin ring. (D) When FUS had the NES deleted (FUS ΔNES), FUS protein predominantly localized inside the nucleus similar to the FUS WT alone [compare (B) with (D)]. (E) The NES double-mutant (FUS- ΔNES R518K) was also retained in the nucleus similar to the FUS WT and FUS ΔNES. Genotypes used here are OK371-GAL4/+, OK371-GAL4;UAS-FUS WT, OK371-GAL4;UAS-FUS R18K, OK371-GAL4;UAS-FUS ΔNES and OK371-GAL4;UAS-FUS ΔNES R518K.
Figure 6.
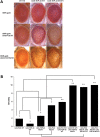
Ectopic expression of WT FUS/TLS enhances neurodegeneration associated with the disease-causing TDP-43 mutation M337V. (A) Light micrographs of Day 1 adult eyes expressing GMR-GAL4/+, TDP-43 or TDP-43 M337V (top panel). The eyes of control flies (GMR-GAL4/+) and flies expressing TDP-43 WT had almost normal and organized ommatidial architecture. The eyes of flies expressing mutant TDP-43 M337V showed mild eye degeneration as reported previously (25). Co-expression of FUS WT and TDP-43 WT caused a rough eye phenotype with ommatidial degeneration, and ommatidial fusion suggesting a synergistic enhancement in the eye degeneration. In addition, co-expression of FUS WT and TDP-43 M337V caused a severe rough eye phenotype (much rougher when compared with the flies co-expressing FUS WT and TDP-43 WT). (B) Quantitative analysis of eye phenotypes. Flies from each genotype were randomly selected for objective scoring according to the criteria described in Materials and Methods. It was clear that coexpression of WT FUS with mutated TDP-43 M337V or co-expression of mutated FUS R521H with WT TDP-43 resulted in a much more severe phenotype than co-expression of WT FUS and WT TDP-43 (based on a scale of 1–10 where 10 is the maximum score). Values are the means ± SEM. The dendrograms illustrate the significant differences between the two genotypes used for the respective crosses (*) with each genotype being significantly different from the other genotype in eye degeneration severity (P < 0.01 in all cases). It also illustrates the significant difference between the resulting cross (**) with each of the genotypes (*) used for the respective cross (P < 0.01 in all cases).
Similar articles
- RNA-binding ability of FUS regulates neurodegeneration, cytoplasmic mislocalization and incorporation into stress granules associated with FUS carrying ALS-linked mutations.
Daigle JG, Lanson NA Jr, Smith RB, Casci I, Maltare A, Monaghan J, Nichols CD, Kryndushkin D, Shewmaker F, Pandey UB. Daigle JG, et al. Hum Mol Genet. 2013 Mar 15;22(6):1193-205. doi: 10.1093/hmg/dds526. Epub 2012 Dec 20. Hum Mol Genet. 2013. PMID: 23257289 Free PMC article. - Inactivation of Hippo and cJun-N-terminal Kinase (JNK) signaling mitigate FUS mediated neurodegeneration in vivo.
Gogia N, Sarkar A, Mehta AS, Ramesh N, Deshpande P, Kango-Singh M, Pandey UB, Singh A. Gogia N, et al. Neurobiol Dis. 2020 Jul;140:104837. doi: 10.1016/j.nbd.2020.104837. Epub 2020 Mar 19. Neurobiol Dis. 2020. PMID: 32199908 Free PMC article. - Conjoint pathologic cascades mediated by ALS/FTLD-U linked RNA-binding proteins TDP-43 and FUS.
Ito D, Suzuki N. Ito D, et al. Neurology. 2011 Oct 25;77(17):1636-43. doi: 10.1212/WNL.0b013e3182343365. Epub 2011 Sep 28. Neurology. 2011. PMID: 21956718 Free PMC article. Review. - Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs.
Lagier-Tourenne C, Polymenidou M, Hutt KR, Vu AQ, Baughn M, Huelga SC, Clutario KM, Ling SC, Liang TY, Mazur C, Wancewicz E, Kim AS, Watt A, Freier S, Hicks GG, Donohue JP, Shiue L, Bennett CF, Ravits J, Cleveland DW, Yeo GW. Lagier-Tourenne C, et al. Nat Neurosci. 2012 Nov;15(11):1488-97. doi: 10.1038/nn.3230. Epub 2012 Sep 30. Nat Neurosci. 2012. PMID: 23023293 Free PMC article. - TDP-43 and FUS en route from the nucleus to the cytoplasm.
Ederle H, Dormann D. Ederle H, et al. FEBS Lett. 2017 Jun;591(11):1489-1507. doi: 10.1002/1873-3468.12646. Epub 2017 Apr 23. FEBS Lett. 2017. PMID: 28380257 Review.
Cited by
- Model systems of motor neuron diseases as a platform for studying pathogenic mechanisms and searching for therapeutic agents.
Valetdinova KR, Medvedev SP, Zakian SM. Valetdinova KR, et al. Acta Naturae. 2015 Jan-Mar;7(1):19-36. Acta Naturae. 2015. PMID: 25926999 Free PMC article. - RNA Granules and Diseases: A Case Study of Stress Granules in ALS and FTLD.
Fan AC, Leung AK. Fan AC, et al. Adv Exp Med Biol. 2016;907:263-96. doi: 10.1007/978-3-319-29073-7_11. Adv Exp Med Biol. 2016. PMID: 27256390 Free PMC article. - DDX17 is involved in DNA damage repair and modifies FUS toxicity in an RGG-domain dependent manner.
Fortuna TR, Kour S, Anderson EN, Ward C, Rajasundaram D, Donnelly CJ, Hermann A, Wyne H, Shewmaker F, Pandey UB. Fortuna TR, et al. Acta Neuropathol. 2021 Sep;142(3):515-536. doi: 10.1007/s00401-021-02333-z. Epub 2021 Jun 1. Acta Neuropathol. 2021. PMID: 34061233 Free PMC article. - Expression of Fused in sarcoma mutations in mice recapitulates the neuropathology of FUS proteinopathies and provides insight into disease pathogenesis.
Verbeeck C, Deng Q, Dejesus-Hernandez M, Taylor G, Ceballos-Diaz C, Kocerha J, Golde T, Das P, Rademakers R, Dickson DW, Kukar T. Verbeeck C, et al. Mol Neurodegener. 2012 Oct 10;7:53. doi: 10.1186/1750-1326-7-53. Mol Neurodegener. 2012. PMID: 23046583 Free PMC article. - Neurodegeneration the RNA way.
Renoux AJ, Todd PK. Renoux AJ, et al. Prog Neurobiol. 2012 May;97(2):173-89. doi: 10.1016/j.pneurobio.2011.10.006. Epub 2011 Nov 3. Prog Neurobiol. 2012. PMID: 22079416 Free PMC article. Review.
References
- Kabashi E., Valdmanis P.N., Dion P., Spielgelman D., McConkey B.J., Vande Velde C., Bouchard J.P., Lacomblez L., Pochigaeva K., Salachas F., et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat. Genet. 2008;40:572–574. doi: 10.1038/ng.132. - DOI - PubMed
- Kwiatkowski T.J., Jr., Bosco D.A., Leclerc A.L., Tamrazian E., Vanderburg C.R., Russ C., Davis A., Gilchrist J., Kasarskis E.J., Munsat T., et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. - DOI - PubMed
- Vance C., Rogelj B., Hortobágyi T., De Vos K.J., Nishimura A.L., Sreedharan J., Hu X., Smith B., Ruddy D., Wright P., et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous