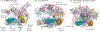Unravelling the means to an end: RNA polymerase II transcription termination - PubMed (original) (raw)
Review
Unravelling the means to an end: RNA polymerase II transcription termination
Jason N Kuehner et al. Nat Rev Mol Cell Biol. 2011 May.
Abstract
The pervasiveness of RNA synthesis in eukaryotes is largely the result of RNA polymerase II (Pol II)-mediated transcription, and termination of its activity is necessary to partition the genome and maintain the proper expression of neighbouring genes. Despite its ever-increasing biological significance, transcription termination remains one of the least understood processes in gene expression. However, recent mechanistic studies have revealed a striking convergence among several overlapping models of termination, including the poly(A)- and Sen1-dependent pathways, as well as new insights into the specificity of Pol II termination among its diverse gene targets. Broader knowledge of the role of Pol II carboxy-terminal domain phosphorylation in promoting alternative mechanisms of termination has also been gained.
Figures
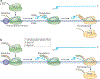
Figure 1 |. Pol II transcription is coordinated with distinct patterns of C‑terminal domain phosphorylation.
a | Initiation: RNA polymerase II (Pol II) is recruited to a gene promoter by transcription factors, the DNA is melted to expose the template strand, and the first few nucleotides of RNA are synthesized. Elongation: a full-length RNA–DNA hybrid is formed (~8–9 bp) and Pol II proceeds to extend the transcript. Termination: Pol II ceases RNA synthesis and becomes termination-prone (indicated by its change in colour from green to yellow), and both Pol II and the nascent RNA are released from the template. As shown, destabilization of the RNA–DNA hybrid in the Pol II active site is likely to be a key feature in termination. Protein factors involved in elongation, RNA processing and termination (shown as yellow, blue and orange ovals, respectively) co-transcriptionally associate with the Pol II carboxy‑terminal domain (CTD). b | The phosphorylation status of the CTD heptad repeats (Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7) changes as Pol II progresses through a gene,. Hypophosphorylated Pol II is recruited into the pre-initiation complexes and it is phosphorylated on Ser5 by the transcription initiation factor IIH (TFIIH) kinase Kin28 (cyclin-dependent kinase 7 (CDK7) in mammals) during initiation and on Ser2 by CTD kinase subunit 1 (Ctk1; CDK9 in mammals) during elongation. The action of these kinases, combined with their opposing phosphatases (suppressor of Sua7 2 (Ssu72) and regulator of transcription 1 (Rtr1) for Ser5‑P and Fcp1 for Ser2‑P), sets up a dual gradient of CTD modification, with Ser5‑P and Ser2‑P being more prevalent towards the 5′ end and the 3′ end of the gene, respectively. Several other kinases and phosphatases contribute to this gradient but are not discussed for simplicity. Each phosphorylation shown on the CTD represents a single heptad repeat (not all repeats are shown) with the general phosphorylation patterns indicated.
Figure 2 |. Factors involved in poly(A)-dependent and Sen1-dependent termination.
Counterparts of termination factors in yeast and humans are shown in the same colour, and known interactions between RNA, RNA polymerase II (Pol II) and other factors are indicated by direct contacts. Pol II carboxy-terminal domain (CTD) phosphorylation dynamics are indicated as in FIG. 1b, with Ser2-P being higher than Ser5-P in regions of poly(A)-dependent termination, and the reverse pattern being observed in regions of Sen1-dependent termination. a | In poly(A)-dependent termination in yeast, the 5′–3′ exoribonuclease RNA-trafficking protein 1 (Rat1; XRN2 in mammals) is recruited to Pol II via proteins that interact with phosphorylated Ser2 in the CTD (such as regulator of Ty1 transposition 103 (Rtt103)) and poly(A) site RNA elements (such as the indicated A-rich and U-rich sequences). Rat1 degrades the downstream RNA (dashed light blue line) that results from the 3′-end processing cleavage event (scissors), which may result in disruption of the Pol II active site hybrid that is representative of the ‘torpedo’ model. In addition to contacting the CTD, cleavage and polyadenylation factor (CPF; homologous to human cleavage and polyadenylation specificity factor (CPSF)) may also interact with the body of Pol II through its suppressor of Sua7 2 (Ssu72) subunit. Optimal association of Rat1 with chromatin requires cleavage factor IA (CFIA; homologous to human cleavage stimulatory factor (CstF)), but direct contacts with CFIA have not been reported. b | In Sen1-dependent termination in yeast, the mechanism that applies to most non-coding RNAs, Sen1 is recruited to Pol II via proteins that interact with Pol II Ser5-P CTD (such as Nrd1) and specific RNA elements (such as GUAA repeats). Sen1 may unwind the Pol II active site hybrid via its helicase activity. c | In poly(A)-dependent termination in humans, pausing of human Pol II is induced when CPSF bound to the body of Pol II recognizes the AAUAAA signal sequence that emerges in the nascent transcript (step 1). Upon exposure of the GU-rich binding site, CstF dislodges CPSF (step 2). Following cleavage at the poly(A) site, 5′–3′ exoribonuclease 2 (XRN2) degrades the downstream RNA product, which may displace Pol II as described above for Rat1 (step 3). CFIIm, mammalian CFII; DOM3Z, DOM-3 homologue Z; Nab3, nuclear polyadenylated RNA-binding 3; Rai1, Rat1-interacting 1.
Figure 3 |. Comparison of E. coli RNA polymerase and S. cerevisiae Pol II structures.
A summary of the putative sites on bacterial RNA polymerase that are contacted by Rho, N utilization substance A (NusA) and NusG, and their corresponding regions in yeast RNA polymerase II (Pol II), are shown. a,b | A molecular model of Escherichia coli RNA polymerase (a) and a crystal structure model of Saccharomyces cerevisiae Pol II (b) are depicted in cartoon rendering (β′-subunit (Rpb1 in S. cerevisiae), pale teal; β-subunit (Rpb2 in S. cerevisiae), light pink; other subunits, wheat; active site, red). The key sites of interaction on E. coli RNA polymerase are highlighted in space-filled rendering (β-flap, yellow; β′-clamp helices, teal; β′-lid, violet; β′-dock, orange), as are the relative sites on Pol II. Note that only the flanking amino-terminal and carboxy-terminal residues of the Pol II Rpb2 flap are shown as the loop is too disordered to be depicted here. c | Crystal structure model of the S. cerevisiae Pol II elongation complex depicted as detailed for E. coli RNA polymerase in (a), but rotated to show the trigger loop (highlighted in blue in space-filled rendering). The template DNA (green) and the nascent transcript (red) are also shown, to emphasize the close proximity of the features detailed in (a) to the RNA exit path. For simplicity, the Rpb4 and Rpb7 subunits of the Pol II elongation complex are hidden. The structures in this figure were created with
PyMOL
(Schrödinger) using
Protein Data Bank
files
3LU0
for RNA polymerase and
1I3Q
and
1Y1W
for Pol II.
Figure 4 |. Genome-wide localization of yeast Pol II CTD phosphorylations and termination factors.
The localization of RNA polymerase II (Pol II) phospho-carboxy-terminal domain (CTD) isoforms (left) and protein 1 of CFI (Pcf11), RNA-trafficking protein 1 (Rat1) and Nrd1 (right) were mapped across the entire yeast genome by ChlP–chip. Average distributions for highly expressed long mRNA-encoding genes (>2,000 bases, 128 genes) (a) and short small nucleolar RNA (snoRNA)-encoding genes (<700 bases, 31 genes) (b) are shown. Transcription units for mRNA-encoding genes (transcription start site (TSS) to poly(A) site) and snoRNA-encoding genes (TSS to mature 3′ end), indicated by arrows, are divided into 10 equal intervals and flanked with 1 kb of 5′ and 3′ sequence. Images are modified, with permission, from REF. 24.
Similar articles
- EM visualization of transcription by RNA polymerase II: downstream termination requires a poly(A) signal but not transcript cleavage.
Osheim YN, Proudfoot NJ, Beyer AL. Osheim YN, et al. Mol Cell. 1999 Mar;3(3):379-87. doi: 10.1016/s1097-2765(00)80465-7. Mol Cell. 1999. PMID: 10198640 - RNA Polymerase II Transcription Attenuation at the Yeast DNA Repair Gene, DEF1, Involves Sen1-Dependent and Polyadenylation Site-Dependent Termination.
Whalen C, Tuohy C, Tallo T, Kaufman JW, Moore C, Kuehner JN. Whalen C, et al. G3 (Bethesda). 2018 May 31;8(6):2043-2058. doi: 10.1534/g3.118.200072. G3 (Bethesda). 2018. PMID: 29686108 Free PMC article. - Effects of Transcription Elongation Rate and Xrn2 Exonuclease Activity on RNA Polymerase II Termination Suggest Widespread Kinetic Competition.
Fong N, Brannan K, Erickson B, Kim H, Cortazar MA, Sheridan RM, Nguyen T, Karp S, Bentley DL. Fong N, et al. Mol Cell. 2015 Oct 15;60(2):256-67. doi: 10.1016/j.molcel.2015.09.026. Mol Cell. 2015. PMID: 26474067 Free PMC article. - Overlapping pathways dictate termination of RNA polymerase II transcription.
Lykke-Andersen S, Jensen TH. Lykke-Andersen S, et al. Biochimie. 2007 Oct;89(10):1177-82. doi: 10.1016/j.biochi.2007.05.007. Epub 2007 Jun 2. Biochimie. 2007. PMID: 17629387 Review. - [Molecular mechanisms of coupling processes of transcription termination and polyadenylation of pro-mRNA].
Zarudnaia MI, Potiagaĭ AL, Dzerzhinskiĭ NE, Gororun DN. Zarudnaia MI, et al. Ukr Biokhim Zh (1999). 2001 Mar-Apr;73(2):28-32. Ukr Biokhim Zh (1999). 2001. PMID: 11642040 Review. Russian.
Cited by
- Swiss army knives: non-canonical functions of nuclear Drosha and Dicer.
Burger K, Gullerova M. Burger K, et al. Nat Rev Mol Cell Biol. 2015 Jul;16(7):417-30. doi: 10.1038/nrm3994. Epub 2015 May 28. Nat Rev Mol Cell Biol. 2015. PMID: 26016561 Review. - The Integrator complex controls the termination of transcription at diverse classes of gene targets.
Skaar JR, Ferris AL, Wu X, Saraf A, Khanna KK, Florens L, Washburn MP, Hughes SH, Pagano M. Skaar JR, et al. Cell Res. 2015 Mar;25(3):288-305. doi: 10.1038/cr.2015.19. Epub 2015 Feb 13. Cell Res. 2015. PMID: 25675981 Free PMC article. - The Reb1-homologue Ydr026c/Nsi1 is required for efficient RNA polymerase I termination in yeast.
Reiter A, Hamperl S, Seitz H, Merkl P, Perez-Fernandez J, Williams L, Gerber J, Németh A, Léger I, Gadal O, Milkereit P, Griesenbeck J, Tschochner H. Reiter A, et al. EMBO J. 2012 Aug 15;31(16):3480-93. doi: 10.1038/emboj.2012.185. Epub 2012 Jul 17. EMBO J. 2012. PMID: 22805593 Free PMC article. - R-loop formation at Snord116 mediates topotecan inhibition of Ube3a-antisense and allele-specific chromatin decondensation.
Powell WT, Coulson RL, Gonzales ML, Crary FK, Wong SS, Adams S, Ach RA, Tsang P, Yamada NA, Yasui DH, Chédin F, LaSalle JM. Powell WT, et al. Proc Natl Acad Sci U S A. 2013 Aug 20;110(34):13938-43. doi: 10.1073/pnas.1305426110. Epub 2013 Aug 5. Proc Natl Acad Sci U S A. 2013. PMID: 23918391 Free PMC article. - Reaction Mechanisms of Pol IV, RDR2, and DCL3 Drive RNA Channeling in the siRNA-Directed DNA Methylation Pathway.
Singh J, Mishra V, Wang F, Huang HY, Pikaard CS. Singh J, et al. Mol Cell. 2019 Aug 8;75(3):576-589.e5. doi: 10.1016/j.molcel.2019.07.008. Mol Cell. 2019. PMID: 31398324 Free PMC article.
References
- Lee TI & Young RA Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet 34, 77–137 (2000). - PubMed
- Espinosa JM The meaning of pausing. Mol. Cell 40, 507–508 (2010). - PubMed
- Rosonina E, Kaneko S. & Manley JL Terminating the transcript: breaking up is hard to do. Genes Dev. 20, 1050–1056 (2006). - PubMed
- Gilmour DS & Fan R. Derailing the locomotive: transcription termination. J. Biol. Chem 283, 661–664 (2008). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K12GM074869/GM/NIGMS NIH HHS/United States
- R01 GM041752/GM/NIGMS NIH HHS/United States
- R01GM041752/GM/NIGMS NIH HHS/United States
- K12 GM074869/GM/NIGMS NIH HHS/United States
- R01GM068887/GM/NIGMS NIH HHS/United States
- R01 GM068887/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources