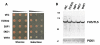A yeast model of FUS/TLS-dependent cytotoxicity - PubMed (original) (raw)
. 2011 Apr;9(4):e1001052.
doi: 10.1371/journal.pbio.1001052. Epub 2011 Apr 26.
Daniel F Tardiff, Haesun Han, Kanneganti Divya, Quan Zhong, Lynne E Maquat, Daryl A Bosco, Lawrence J Hayward, Robert H Brown Jr, Susan Lindquist, Dagmar Ringe, Gregory A Petsko
Affiliations
- PMID: 21541368
- PMCID: PMC3082520
- DOI: 10.1371/journal.pbio.1001052
A yeast model of FUS/TLS-dependent cytotoxicity
Shulin Ju et al. PLoS Biol. 2011 Apr.
Abstract
FUS/TLS is a nucleic acid binding protein that, when mutated, can cause a subset of familial amyotrophic lateral sclerosis (fALS). Although FUS/TLS is normally located predominantly in the nucleus, the pathogenic mutant forms of FUS/TLS traffic to, and form inclusions in, the cytoplasm of affected spinal motor neurons or glia. Here we report a yeast model of human FUS/TLS expression that recapitulates multiple salient features of the pathology of the disease-causing mutant proteins, including nuclear to cytoplasmic translocation, inclusion formation, and cytotoxicity. Protein domain analysis indicates that the carboxyl-terminus of FUS/TLS, where most of the ALS-associated mutations are clustered, is required but not sufficient for the toxicity of the protein. A genome-wide genetic screen using a yeast over-expression library identified five yeast DNA/RNA binding proteins, encoded by the yeast genes ECM32, NAM8, SBP1, SKO1, and VHR1, that rescue the toxicity of human FUS/TLS without changing its expression level, cytoplasmic translocation, or inclusion formation. Furthermore, hUPF1, a human homologue of ECM32, also rescues the toxicity of FUS/TLS in this model, validating the yeast model and implicating a possible insufficiency in RNA processing or the RNA quality control machinery in the mechanism of FUS/TLS mediated toxicity. Examination of the effect of FUS/TLS expression on the decay of selected mRNAs in yeast indicates that the nonsense-mediated decay pathway is probably not the major determinant of either toxicity or suppression.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
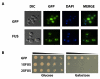
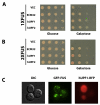
Figure 1. Expression, localization, and toxicity of FUS/TLS in yeast.
(A) Cells expressing GFP or GFP-FUS were induced by 2% galactose for 6 h. Cells were then fixed and viewed by fluorescence microscopy. DAPI was used to stain the nucleus. (B) Yeast with integrated GFP, 1XFUS (1 copy of untagged FUS integrated into the HIS3 locus in the genome), and 2XFUS (2 copies of untagged FUS integrated into the_HIS3_ locus and the TRP1 locus in the genome) were serially diluted (from left to right) and spotted onto plates containing either glucose (FUS expression “off”) or galactose (expression “on”). Picture was taken after 2 d growth at 30°C.
Figure 2. Aggregates of FUS/TLS are different from that of Huntingtin.
(A) Yeast cells containing GFP-tagged N-terminal huntingtin harboring pathogenic polyglutamine expansions (Htt103Q, stretch of 103 consecutive glutamines), normal huntingtin (Htt25Q, stretch of 25 consecutive glutamines), and GFP tagged FUS/TLS were induced with galactose for 6 h. Filter retardation assay was performed to characterize the aggregates. (B) GFP tagged FUS, Htt103Q, or Htt25Q was transformed into_HSP104Δ_, RNQ1Δ deletion strains and the isogenic wild type strain BY4743 (WT). Cells were serially diluted and spotted onto glucose (expression “off”) or galactose plate (expression “on”) to observe toxicity. Pictures were taken after 2 d growth at 30°C. GFP on the same vector (GFP) was used as control. (C) The same strains as above were visualized for localization and aggregation of the proteins by fluorescence microscopy. (D) pYES2CT/GFP-FUS (FUS) and empty vector (Vec) were transformed into wild type yeast, and yeast containing integrated Htt103Q. Freshly grown cells were then serially diluted and spotted onto glucose (expression “off”) or galactose plate (expression “on”) to observe toxicity. Pictures were taken after 2 d growth at 30°C.
Figure 3. Toxicity and localization of individual domains of FUS/TLS.
(A) A serial deletion of the full length FUS/TLS gene from c-terminus, labeled as 1–4, and from N-terminus, labeled as 6–8, was carried out. (B) The truncated genes (with GFP tag at N-terminus) were then placed under the control of GAL1 promoter on the yeast expression vector pYES2CT. Yeast with above constructs was serially diluted and spotted onto plate containing either glucose (expression “off”) or galactose (expression “on”). Pictures of the plates were taken after 2 d growth at 30°C. (C) Cells containing the above constructs were grown in the Ura-Raffinose medium to mid-log phase. Expression of the proteins was induced by 2% galactose for 6 h. Localization and aggregation of the proteins was visualized by fluorescence microscopy. (D) Constructs 3–5 as shown in (A) were also cloned into yeast expression vector pDEST52 without the GFP tag. Yeast containing the constructs was serially diluted and spotted onto glucose (expression “off”) or galactose plate (expression “on”). Picture of the plates was taken after 2 d growth at 30°C.
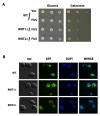
Figure 4. Deletion of arginine methyl transferase does not rescue FUS/TLS toxicity nor change its localization.
(A) Empty vector and N-terminus GFP-tagged FUS/TLS on pYES2CT were transformed into yeast arginine methyl transferase deletion strain_rmt1Δ_, rmt2Δ, and its isogenic wild type BY4743 (WT). Spotting assay was performed to observe toxicity from the above yeast strains. (B) Expression of the proteins from above strains was induced by 2% galactose for 6 h. Localization and aggregation of the protein was visualized by fluorescence microscopy.
Figure 5. FUS_plus and FUS_switch promote nuclear localization and lower toxicity of FUS/TLS.
(A) GFP was fused with the nuclear localization signal of FUS/TLS (GFP_FUS) and Hrp1 (GFP_Hrp1) as shown in top part of (A). Expression of protein from yeast containing above constructs was induced by 2% galactose for 6 h. Localization and aggregation of protein was visualized by fluorescence microscopy. (B) Hrp1 NLS was added to the c-terminal end of FUS/TLS (FUS_plus) or was used to replace nuclear localization signal of FUS/TLS (FUS_switch), as shown on part top of (B). Protein expression from yeast containing above constructs was induced by 0.1% galactose for 6 h. Protein localization and aggregation was visualized by fluorescence microscopy. (C) The same yeast strains were grown in raffinose medium and 0.1% galactose medium. Cell growth was monitored using a Bioscreen machine for 2 d at 30°C. Typically, at least 10 replicates were done for each.
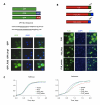
Figure 6. Expression of ECM32, SBP1, SKO1, and VHR1 rescues FUS/TLS toxicity.
(A) ECM32, SBP1, SKO1, and VHR1 were individually transformed into 1X FUS strain. Spotting assay was performed to observe toxicity of yeast containing the above constructs. (B) Protein expression was induced by 2% galactose for 6 h from above yeast strains. Western blot analysis was performed using an antibody against FUS/TLS. PGK1 is shown as a control of protein loading.
Figure 7. hUPF1 rescues FUS/TLS toxicity.
hUPF1 and hUPF2 were cloned into yeast expression vector pYES2CT. (A) The constructs were transformed into 1XFUS (one copy of FUS/TLS integrated at HIS locus). Spotting assay was performed to check the rescue of toxicity by hUPF1 and_hUPF2._ Empty vector and ECM32_construct from library screen were used as negative and positive controls. (B) The above constructs were transformed into 2XFUS (two copies of FUS/TLS integrated at HIS locus and TRP locus, respectively). Spotting assay was performed to check the rescue of toxicity by_hUPF1 and hUPF2. (C) GFP tagged FUS/TLS (pYES2CT/GFP-FUS) and RFP tagged hUPF1 (pRSGal1hUPF1-DsRed) were transformed into yeast. Protein expression was induced by 2% galactose for 6 h and visualized by fluorescence microcopy.
Figure 8. Rescue of FUS/TLS toxicity requires full length hUPF1, and rescue is not mediated by decrease in FUS/TLS protein level or inclusion formation of the protein.
(A) Protein expression was induced by 2% galactose for 6 h in 1XFUS strain expressing hUPF1, hUPF2, or ECM32. Western blot was performed using an antibody against FUS/TLS to check the expression of FUS/TLS. PGK1 is shown as a control of protein loading; (B) the same yeast cells were also subject to indirect immunofluorescence staining using primary antibody against FUS/TLS, and secondary antibody conjugated with fluorescein. Nuclear DNA was stained with DAPI. (C) Different domains of hUPF1 were cloned into a yeast expression vector and transformed into 1XFUS strain. Spotting assay was performed to check rescue of the toxicity. hUPF1-418, hUPF2 binding domain; hUPF419-1118, ATPase/Helicase domain; hUPF1R843C, inactivated ATPase/Helicase by arginine to cysteine mutation at residue of 843 in the full length FUS/TLS.
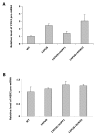
Figure 9. Yeast CYH2 but not MER2 pre-mRNA was accumulated when FUS is over-expressed.
Cells were grown in raffinose medium to early log phase. Protein expression was induced by 2% galactose for 6 h in 1XFUS strain expressing empty vector (1XFUS), human UPF1 (+hUPF1), or_ECM32_ (+ECM32). CYH2 pre-mRNA (A) and MER2 pre-mRNA level (B) were determined by qRT-PCR using 18sRNA as an internal control. Pre-mRNA in wild type yeast cell without integration of FUS (WT) was normalized to 1.
Comment in
- A yeast model for understanding ALS: fast, cheap, and easy to control.
Robinson R. Robinson R. PLoS Biol. 2011 Apr;9(4):e1001053. doi: 10.1371/journal.pbio.1001053. Epub 2011 Apr 26. PLoS Biol. 2011. PMID: 21541366 Free PMC article. No abstract available.
Similar articles
- Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS.
Sun Z, Diaz Z, Fang X, Hart MP, Chesi A, Shorter J, Gitler AD. Sun Z, et al. PLoS Biol. 2011 Apr;9(4):e1000614. doi: 10.1371/journal.pbio.1000614. Epub 2011 Apr 26. PLoS Biol. 2011. PMID: 21541367 Free PMC article. - A Genetic Screen for Human Genes Suppressing FUS Induced Toxicity in Yeast.
Hayden E, Chen S, Chumley A, Xia C, Zhong Q, Ju S. Hayden E, et al. G3 (Bethesda). 2020 Jun 1;10(6):1843-1852. doi: 10.1534/g3.120.401164. G3 (Bethesda). 2020. PMID: 32276960 Free PMC article. - FUS/TLS forms cytoplasmic aggregates, inhibits cell growth and interacts with TDP-43 in a yeast model of amyotrophic lateral sclerosis.
Kryndushkin D, Wickner RB, Shewmaker F. Kryndushkin D, et al. Protein Cell. 2011 Mar;2(3):223-36. doi: 10.1007/s13238-011-1525-0. Epub 2011 Mar 30. Protein Cell. 2011. PMID: 21452073 Free PMC article. - TDP-43 and FUS/TLS: sending a complex message about messenger RNA in amyotrophic lateral sclerosis?
Strong MJ, Volkening K. Strong MJ, et al. FEBS J. 2011 Oct;278(19):3569-77. doi: 10.1111/j.1742-4658.2011.08277.x. Epub 2011 Sep 6. FEBS J. 2011. PMID: 21810174 Review. - Understanding the role of TDP-43 and FUS/TLS in ALS and beyond.
Da Cruz S, Cleveland DW. Da Cruz S, et al. Curr Opin Neurobiol. 2011 Dec;21(6):904-19. doi: 10.1016/j.conb.2011.05.029. Epub 2011 Aug 2. Curr Opin Neurobiol. 2011. PMID: 21813273 Free PMC article. Review.
Cited by
- Mutant TDP-43 and FUS cause age-dependent paralysis and neurodegeneration in C. elegans.
Vaccaro A, Tauffenberger A, Aggad D, Rouleau G, Drapeau P, Parker JA. Vaccaro A, et al. PLoS One. 2012;7(2):e31321. doi: 10.1371/journal.pone.0031321. Epub 2012 Feb 21. PLoS One. 2012. PMID: 22363618 Free PMC article. - Latrepirdine stimulates autophagy and reduces accumulation of α-synuclein in cells and in mouse brain.
Steele JW, Ju S, Lachenmayer ML, Liken J, Stock A, Kim SH, Delgado LM, Alfaro IE, Bernales S, Verdile G, Bharadwaj P, Gupta V, Barr R, Friss A, Dolios G, Wang R, Ringe D, Protter AA, Martins RN, Ehrlich ME, Yue Z, Petsko GA, Gandy S. Steele JW, et al. Mol Psychiatry. 2013 Aug;18(8):882-8. doi: 10.1038/mp.2012.115. Epub 2012 Aug 7. Mol Psychiatry. 2013. PMID: 22869031 Free PMC article. - Characterization of FUS mutations in amyotrophic lateral sclerosis using RNA-Seq.
van Blitterswijk M, Wang ET, Friedman BA, Keagle PJ, Lowe P, Leclerc AL, van den Berg LH, Housman DE, Veldink JH, Landers JE. van Blitterswijk M, et al. PLoS One. 2013;8(4):e60788. doi: 10.1371/journal.pone.0060788. Epub 2013 Apr 8. PLoS One. 2013. PMID: 23577159 Free PMC article. - TDP-43 Toxicity in Yeast Is Associated with a Reduction in Autophagy, and Deletions of TIP41 and PBP1 Counteract These Effects.
Park SK, Park S, Liebman SW. Park SK, et al. Viruses. 2022 Oct 15;14(10):2264. doi: 10.3390/v14102264. Viruses. 2022. PMID: 36298819 Free PMC article. - TDP43 and RNA instability in amyotrophic lateral sclerosis.
Weskamp K, Barmada SJ. Weskamp K, et al. Brain Res. 2018 Aug 15;1693(Pt A):67-74. doi: 10.1016/j.brainres.2018.01.015. Epub 2018 Jan 31. Brain Res. 2018. PMID: 29395044 Free PMC article. Review.
References
- Rosen D. R, Siddique T, Patterson D, Figlewicz D. A, Sapp P, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. - PubMed
- Kwiatkowski T. J, Jr, Bosco D. A, Leclerc A. L, Tamrazian E, Vanderburg C. R, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- RC2 NS070342/NS/NINDS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- 1RC1NS06839/NS/NINDS NIH HHS/United States
- NS614192/NS/NINDS NIH HHS/United States
- RC1 NS068391/NS/NINDS NIH HHS/United States
- U01NS05225-03/NS/NINDS NIH HHS/United States
- R01NS050557-05/NS/NINDS NIH HHS/United States
- R01 NS050557/NS/NINDS NIH HHS/United States
- 1RC2NS070342-01/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous