Differential curvature sensing and generating activities of dynamin isoforms provide opportunities for tissue-specific regulation - PubMed (original) (raw)
Differential curvature sensing and generating activities of dynamin isoforms provide opportunities for tissue-specific regulation
Ya-Wen Liu et al. Proc Natl Acad Sci U S A. 2011.
Abstract
Dynamin 1 (Dyn1) and Dyn2 are neuronal and ubiquitously expressed isoforms, respectively, of the multidomain GTPase required for clathrin-mediated endocytosis (CME). Although they are 79% identical, Dyn1 and Dyn2 are not fully functionally redundant. Through direct measurements of basal and assembly-stimulated GTPase activities, membrane binding, self-assembly, and membrane fission on planar and curved templates, we have shown that Dyn1 is an efficient curvature generator, whereas Dyn2 is primarily a curvature sensor. Using Dyn1/Dyn2 chimeras, we identified the lipid-binding pleckstrin homology domain as being responsible for the differential in vitro properties of these two isoforms. Remarkably, their in vitro activities were reversed by a single amino acid change in the membrane-binding variable loop 3. Reconstitution of KO mouse embryo fibroblasts showed that both the pleckstrin homology and the Pro/Arg-rich domains determine the differential abilities of these two isoforms to support CME. These domains are specific to classical dynamins and are involved in regulating their activity. Our findings reveal opportunities for fundamental differences in the regulation of Dyn1, which mediates rapid endocytosis at the synapse, vs. Dyn2, which regulates early and late events in CME in nonneuronal cells.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
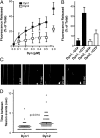
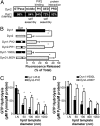
Differential ability of Dyn1 and Dyn2 to release vesicles from planar membranes. (A) The indicated concentrations of Dyn1 or Dyn2 were incubated with SUPER templates for 30 min at room temperature. Membrane fission was measured by the release of fluorescently labeled vesicles into the supernatant after sedimentation of the SUPER templates (shown are averages ± SD, n = 5). (B) Average fission activity of 0.5 μM Dyn1 or Dyn2 (n ≥ 21). (C) Time-lapse images showing fission activity of Dyn2 on membrane tethers drawn from SUPER templates and imaged in the presence of an oxygen scavenger system. Dyn2 was added at a final concentration of 0.5 μM, and images were taken in 1.4-s intervals (
Movie S1
). (D) Quantification of fission activity of Dyn1 and Dyn2 with membrane tethers as templates. Dyn1 or Dyn2 was added at a final concentration of 0.5 μM, and movies were taken with 0.866-s time-lapse intervals. In an attempt to quantify fission activity, the time between fission events on an individual tether was determined. Data are presented as a scattered plot and were analyzed for significance with a Mann–Whitney Test using PRISM (Graphpad) statistical software.
Fig. 2.
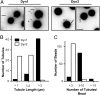
Differential ability of Dyn1 and Dyn2 to generate curvature from planar membranes. (A) Tubulation of SUPER templates. Dyn1 or Dyn2 (0.5 μM) were incubated with SUPER templates for 10 min at room temperature in the absence of GTP and imaged in the presence of an oxygen scavenger system. Images are inverted in contrast for clarity, and arrows indicate tubules. (Scale bar: 5 μm.) (B) Quantification of tubule length. Length of tubules was determined using ImageJ (National Institutes of Health; n > 50 tubules). (C) Quantification of number of tubules per bead (n > 100 beads).
Fig. 3.
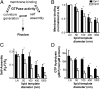
Differential curvature sensitivity of Dyn1 and Dyn2 on lipid templates. (A) The assembly-stimulated GTPase activity of dynamin requires its ability to bind membranes, self-assemble, and generate curvature. Although these activities are interdependent, each can be directly measured. All four of these activities combine and are required for dynamin-catalyzed membrane fission. (B) Membrane binding activity of Dyn1 and Dyn2. Dyn1-G532C-NBD and Dyn2-S532C-NBD were mixed with WT Dyn1 and Dyn2 at a ratio of 1:10 and incubated at room temperature with lipid templates of different curvature (LN, lipid nanotubes, estimated at 30 nm diameter) for 10 min to achieve steady-state binding in the absence of nucleotide. The fold change of fluorescence at 530 nm was measured and normalized to the fold change observed with lipid nanotubes. Data are shown as averages ± SD (n = 9). (C) Self-assembly of Dyn1 and Dyn2 on lipid templates for 10 min to achieve steady-state assembly. Dyn1-BODIPY and Dyn2-S702C-BODIPY were incubated with lipid templates of different curvature in the absence of nucleotide at room temperature, and the assembly-dependent quenching of BODIPY fluorescence was monitored at 510 nm. Data are shown as averages ± SD (n = 5). (D) Lipid-stimulated GTPase activity of Dyn1 and Dyn2. Dynamin (0.5 μM) was incubated with lipid templates of different curvature in the presence of 1 mM GTP at 37 °C. Released Pi was determined using a colorimetric malachite green assay. Data are shown as averages ± SD (n = 4). *P ≤ 0.01
Fig. 4.
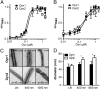
Differential abilities of Dyn1 and Dyn2 to self-assemble and generate curvature on lipid templates. Concentration dependence and cooperative behavior of Dyn1 and Dyn2 GTPase activity on (A) nanotubes or (B) 100-nm liposomes are shown. Data (average ± SD, n = 3) are normalized to maximum activity. (C) Electron micrographs of Dyn1 and Dyn2 assembled onto lipid nanotubes (LN) or 400- or 1,000-nm liposomes as indicated. Dynamin was incubated with lipid templates for 20 min at room temperature, and then, it was adsorbed to a grid and imaged by negative-stain EM. Diameter (d) is in nanometers. (Scale bar: 50 nm.) (D) Quantification of diameter of dynamin-decorated lipid templates (n ≥ 4).
Fig. 5.
A tyrosine residue in the PH domain VL3 confers isoform-specific curvature sensitivity and membrane fission activity. (A) Domain structure of Dyn1 and Dyn2 and sequence identity between both isoforms. Dyn1-PH2 and Dyn2-PH1 chimeras, as illustrated in B, were generated by seamless cloning (
Fig. S2_A_
). (B) Fission activity of parent chimeric Dyn1 and Dyn2 or single amino acid-substituted Dyn1 and Dyn2 (0.5 μM), as indicated, was determined after incubation with SUPER templates by sedimentation and release of fluorescent vesicles into the supernatant. Data shown are averages ± SD (n ≥ 16, *P ≤ 0.001). Fluorescence released in the absence of GTP was subtracted as a background. (C and D) Stimulated GTPase activity of chimeric dynamins (C) or single amino acid substitution at position 600 in VL3 (D) is shown. Dynamins (0.5 μM) were incubated with lipid templates of different curvature, and released Pi was determined using a colorimetric malachite green assay. Data are shown as averages ± SD (n = 3, *P < 0.02).
Fig. 6.
The PH domain and the PRD confer isoform-specific activities in vivo. (A) Conditional Dyn1/Dyn2 null mouse embryo fibroblasts were treated with 4-OHT for 4 d to induce Cre recombinase for KO of endogenous dynamins. Control cells are Dyn1/2-conditional null MEFs reconstituted with retroviruses expressing GFP only and not treated with 4-OHT. Western blot analysis of Dyn2 expression levels before (Control) and after (KO) 4-OHT treatment. Different amounts of cell lysates (1×, 2×, 5×, 10×, 20×, and 50× from left to right per cell line) were loaded and probed using an antibody against Dyn2. A representative blot is shown, and actin serves as a loading control. (B) CME activity was measured by internalization of b-Tfn for 5 min at 37 °C as determined by ELISA and is expressed relative to total surface bound. Data shown are average ± SD (n = 8, *P < 0.05). (C) Western blot analysis of the expression of HA-tagged dynamin proteins in conditional null MEFs after KO of endogenous dynamins. For quantification, see Table 1. Loading control is a background band stained by the HA antibody. (Left) Western blot of control cells and HA-Dyn2 reconstituted cells (different amounts of cell lysate are loaded in each lane). (Right) Western blot of cells reconstituted with indicated dynamins (same amounts of cell lysate are loaded in each lane). (D) Clathrin-mediated endocytosis in control or KO cells expressing indicated dynamin constructs. Uptake of b-Tfn was measured as described above and then normalized for different expression levels of the dynamin constructs to obtain a measure of relative Tfn uptake activity. Data are shown as averages ± SD of n ≥ 3 experiments (P < 0.05 compared with Dyn2).
Fig. 7.
Differential activities of dynamin isoforms at the synapse and in fibroblasts provide opportunities for tissue-specific regulation. (A) Dyn1 is the major isoform at the synapse, and it functions during rapid endocytosis after synaptic vesicle release. Phosphorylation of the PRD inhibits Dyn self-assembly and membrane binding (37). On stimulation by an action potential, Ca2+ influx triggers the release of synaptic vesicles docked at the membrane and activates the phosphatase calcineurin, which in turn, dephosphorylates and activates Dyn1 and other endocytic proteins (44). We propose that Dyn1 can support rapid (_t_1/2 = 1–10 s) membrane fission and retrieval of synaptic vesicle membrane components because of its powerful PH domain, potentially with or without the help of coat proteins. In contrast, Dyn2 in fibroblasts (B) supports a more classical (_t_1/2 = 30 to >120 s) form of endocytosis and is regulated by an assembly switch. Dyn2 is recruited to nascent CCPs through their PRDs, where it can regulate CCP maturation and the formation of a deeply invaginated pit by coat and accessory proteins as illustrated. After the neck is sufficiently narrow, additional Dyn2 is recruited to the membrane through its PH domain, where it can mediate fission. Thus, the differential properties of Dyn1 and Dyn2, as curvature generators or sensors, respectively, are critical for their tissue-specific activities.
Fig. P1.
Differential regulation of dynamin isoforms in fibroblasts and at the synapse. (A) In fibroblasts, Dyn2 is recruited to nascent clathrin-coated pits through its proline/arginine domain, where it can regulate clathrin-coated pit maturation and the formation of a deeply invaginated pit by coat and accessory proteins. After the neck is sufficiently narrow, additional Dyn2 is recruited to the membrane through its curvature-sensitive pleckstrin homology domain to mediate fission. (B) In the resting synapse, the proline/arginine domain of Dyn1 is phosphorylated, inhibiting Dyn1 self-assembly and membrane binding. Dephosphorylation occurs rapidly on stimulation, allowing Dyn1 to rapidly self-assemble and mediate membrane fission and retrieval of synaptic vesicle membrane components, perhaps without the help of coat proteins (as illustrated) because of its powerful curvature-generating pleckstrin homology domain.
Similar articles
- Early and nonredundant functions of dynamin isoforms in clathrin-mediated endocytosis.
Bhave M, Mettlen M, Wang X, Schmid SL. Bhave M, et al. Mol Biol Cell. 2020 Aug 15;31(18):2035-2047. doi: 10.1091/mbc.E20-06-0363. Epub 2020 Jun 24. Mol Biol Cell. 2020. PMID: 32579424 Free PMC article. - A noncanonical role for dynamin-1 in regulating early stages of clathrin-mediated endocytosis in non-neuronal cells.
Srinivasan S, Burckhardt CJ, Bhave M, Chen Z, Chen PH, Wang X, Danuser G, Schmid SL. Srinivasan S, et al. PLoS Biol. 2018 Apr 18;16(4):e2005377. doi: 10.1371/journal.pbio.2005377. eCollection 2018 Apr. PLoS Biol. 2018. PMID: 29668686 Free PMC article. - Isoform and splice-variant specific functions of dynamin-2 revealed by analysis of conditional knock-out cells.
Liu YW, Surka MC, Schroeter T, Lukiyanchuk V, Schmid SL. Liu YW, et al. Mol Biol Cell. 2008 Dec;19(12):5347-59. doi: 10.1091/mbc.e08-08-0890. Epub 2008 Oct 15. Mol Biol Cell. 2008. PMID: 18923138 Free PMC article. - Dynamin: functional design of a membrane fission catalyst.
Schmid SL, Frolov VA. Schmid SL, et al. Annu Rev Cell Dev Biol. 2011;27:79-105. doi: 10.1146/annurev-cellbio-100109-104016. Epub 2011 May 18. Annu Rev Cell Dev Biol. 2011. PMID: 21599493 Review. - Structural insights into dynamin-mediated membrane fission.
Faelber K, Held M, Gao S, Posor Y, Haucke V, Noé F, Daumke O. Faelber K, et al. Structure. 2012 Oct 10;20(10):1621-8. doi: 10.1016/j.str.2012.08.028. Structure. 2012. PMID: 23063009 Review.
Cited by
- Analyzing membrane remodeling and fission using supported bilayers with excess membrane reservoir.
Neumann S, Pucadyil TJ, Schmid SL. Neumann S, et al. Nat Protoc. 2013 Jan;8(1):213-22. doi: 10.1038/nprot.2012.152. Epub 2013 Jan 3. Nat Protoc. 2013. PMID: 23288321 Free PMC article. - F-actin Bundle Sedimentation Assay.
Lin SS, Chuang MC, Liu YW. Lin SS, et al. Bio Protoc. 2019 Nov 5;9(21):e3419. doi: 10.21769/BioProtoc.3419. eCollection 2019 Nov 5. Bio Protoc. 2019. PMID: 33654917 Free PMC article. - Flexible pivoting of dynamin pleckstrin homology domain catalyzes fission: insights into molecular degrees of freedom.
Baratam K, Jha K, Srivastava A. Baratam K, et al. Mol Biol Cell. 2021 Jul 1;32(14):1306-1319. doi: 10.1091/mbc.E20-12-0794. Epub 2021 May 12. Mol Biol Cell. 2021. PMID: 33979205 Free PMC article. - Structural Insights into the Mechanism of Dynamin Superfamily Proteins.
Jimah JR, Hinshaw JE. Jimah JR, et al. Trends Cell Biol. 2019 Mar;29(3):257-273. doi: 10.1016/j.tcb.2018.11.003. Epub 2018 Dec 5. Trends Cell Biol. 2019. PMID: 30527453 Free PMC article. Review. - Steric interference from intrinsically disordered regions controls dynamin-related protein 1 self-assembly during mitochondrial fission.
Lu B, Kennedy B, Clinton RW, Wang EJ, McHugh D, Stepanyants N, Macdonald PJ, Mears JA, Qi X, Ramachandran R. Lu B, et al. Sci Rep. 2018 Jul 18;8(1):10879. doi: 10.1038/s41598-018-29001-9. Sci Rep. 2018. PMID: 30022112 Free PMC article.
References
- Ehrlich M, et al. Endocytosis by random initiation and stabilization of clathrin-coated pits. Cell. 2004;118:591–605. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01MH61345/MH/NIMH NIH HHS/United States
- 2R37NS036251/NS/NINDS NIH HHS/United States
- R01 MH061345/MH/NIMH NIH HHS/United States
- R01GM42455/GM/NIGMS NIH HHS/United States
- R01 GM042455/GM/NIGMS NIH HHS/United States
- R37 NS036251/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials