Sorting of GPI-anchored proteins into ER exit sites by p24 proteins is dependent on remodeled GPI - PubMed (original) (raw)
Sorting of GPI-anchored proteins into ER exit sites by p24 proteins is dependent on remodeled GPI
Morihisa Fujita et al. J Cell Biol. 2011.
Abstract
Glycosylphosphatidylinositol (GPI) anchoring of proteins is a posttranslational modification occurring in the endoplasmic reticulum (ER). After GPI attachment, proteins are transported by coat protein complex II (COPII)-coated vesicles from the ER. Because GPI-anchored proteins (GPI-APs) are localized in the lumen, they cannot interact with cytosolic COPII components directly. Receptors that link GPI-APs to COPII are thought to be involved in efficient packaging of GPI-APs into vesicles; however, mechanisms of GPI-AP sorting are not well understood. Here we describe two remodeling reactions for GPI anchors, mediated by PGAP1 and PGAP5, which were required for sorting of GPI-APs to ER exit sites. The p24 family of proteins recognized the remodeled GPI-APs and sorted them into COPII vesicles. Association of p24 proteins with GPI-APs was pH dependent, which suggests that they bind in the ER and dissociate in post-ER acidic compartments. Our results indicate that p24 complexes act as cargo receptors for correctly remodeled GPI-APs to be sorted into COPII vesicles.
Figures
Figure 1.
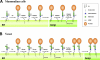
Remodeling of GPI-APs in mammalian cells and yeast. (A) Remodeling of mammalian GPI-APs. Biosynthesis of GPI is performed in the ER through an enzymatic reaction pathway consisting of 10 steps (Kinoshita et al., 2008; Fujita and Kinoshita, 2010). Preformed GPI is attached to the C-terminus of a newly synthesized protein by GPI transamidase (TA). The acyl chain linked to inositol is eliminated by PGAP1. A side-chain EtNP attached to the second mannose is removed by PGAP5. After arrival of GPI-APs at the Golgi, fatty acid remodeling occurs; an unsaturated acid at the sn-2 position in the GPI lipid is removed by PGAP3 and then a saturated fatty acid (stearic acid) is transferred back. PGAP2 is a noncatalytic protein involved in the latter reaction. The putative catalytic component of the acyltransferase (Acyl-T) has yet to be discovered. (B) Remodeling of GPI-APs in the yeast Saccharomyces cerevisiae. After attachment of GPI to proteins, the acyl chain linked to inositol is eliminated by Bst1p (PGAP1 homologue). Then, the fatty acid remodeling of the GPI anchor is performed in the ER, mediated by Per1p (PGAP3 homologue) and Gup1p. In yeast, the diacylglycerol moiety in many GPI-APs is exchanged with ceramide by Cwh43p. Ted1p and Cdc1p, homologues of mammalian PGAP5, are localized in the ER. It was reported that Ted1p acts together with Emp24p and Erv25p, and that Cdc1p functions after Per1p and Gup1p in the same pathway, which suggests that they are involved in GPI remodeling. Side-chain EtNPs on the first and second mannoses are required for ceramide remodeling, whereas it remains unclear whether the side-chain EtNPs are eliminated.
Figure 2.
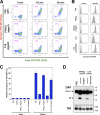
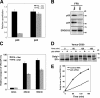
Efficient transport of GPI-APs requires structural remodeling of GPI anchors by PGAP1 and PGAP5. (A) Flow cytometric analysis of transport of GPI-anchored reporter protein. Parental FF8 (WT), pgap1 mutant FPRC2, and pgap5 mutant C19 cells expressing VFG-GPI were stained with an anti-Flag antibody at the indicated times after a temperature shift from 40°C to 32°C. The percentages of cells in the pentagonal region are indicated. (B and C) The cell surface expression of VFG-GPI was analyzed by flow cytometry. FF8 (WT), FPRC2 (pgap1), C19 (pgap5), and FPRC2 cells stably expressing PGAP1 (+ PGAP1) and C19 cells stably expressing HA-tagged PGAP5 (+HA-PGAP5) were stained with anti-Flag antibody at the indicated times after the commencement of transport. The geometric mean fluorescence values of surface VFG-GPI in all cells shown in histograms (B) were quantified at each time point (C). The geometric mean of WT cells at 60 min was plotted as 100% relative transport. Values represent mean ± SD (error bars; n = 3). (D) Western blotting analysis of DAF in the steady state. Cell lysates of FF8 (WT), FPRC2 (pgap1), C19 (pgap5), and FPRC2 cells stably transfected with PGAP1, and C19 cells stably transfected with HA-PGAP5 were analyzed with an anti-DAF antibody under nonreducing conditions to evaluate the steady-state levels of the ER and mature forms. TfR, transferrin receptor for loading assessment.
Figure 3.
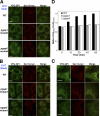
Sorting of GPI-APs into the ERES is impaired in pgap1 and pgap5 mutant cells. (A–C) VFG-GPI were expressed and accumulated in the ER by incubating FF8 (WT), FPRC2 (pgap1), and C19 (pgap5) cells stably expressing myc-tagged Sec13 (Sec13-myc) with 1 µg/ml doxycycline at 40°C for 24 h. After incubation at 10°C for 0 min (A) or 30 min (B and C, high magnification of B), cells were fixed with 4% paraformaldehyde and stained with anti-myc antibody to label the ERES. Bars: (A and B) 10 µm; (C) 5 µm. (D) Relative intensity of VFG-GPI in the ERES. The ratios of average intensities of VFG-GPI in the ERES (AI(eres)) to those in the ERES surroundings (AI(surro)) were measured. The ratios between wild-type (WT) and pgap1 mutant cells and between WT and pgap5 mutant cells differed significantly at each time point. (n = 715–1,906; Student’s t test, P < 0.0001). Error bars indicate the standard error of the mean.
Figure 4.
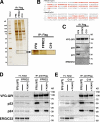
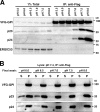
Association of p23 and p24 with remodeled GPI-APs. (A) Detection of proteins specifically associated with VFG-GPI in FF8. VFG-GPI was expressed and accumulated in the ER in FF8, FPRC2, and C19 cells. The cells were then incubated at 32°C for 20 min to initiate transport of VFG-GPI. After cell lysis, VFG-GPI was precipitated with anti-Flag beads. Co-precipitated proteins were eluted using the Flag peptide and subjected to SDS-PAGE and silver staining. The boxed area at ∼20 kD is enlarged to the right. (B) Sequences of hamster Tmed10 (p23) and Tmed2 (p24). Protein bands at 20 kD in A were digested in-gel with trypsin and analyzed by mass spectrometry. The fragments detected by MS/MS analysis are shown in red. (C) Precipitated proteins in A were analyzed by Western blotting using a rabbit anti-p23, anti-p24, or anti-ERGIC53 polyclonal antibody. VFG-GPI was detected with an anti-GFP antibody. (D and E) Immunoprecipitation of VFG-GPI with p23 and p24. FF8, FPRC2, and FPRC2 stably expressing PGAP1 (D) or FF8, C19, and C19 stably expressing HA-PGAP5 (E) were cultured with 1 µg/ml doxycycline at 40°C for 24 h to induce VFG-GPI expression and its accumulation in the ER. The cells were then incubated at 32°C for 20 min to initiate VFG-GPI transport. After cell lysis, VFG-GPI was precipitated with anti-Flag beads and coprecipitated proteins were detected by immunoblotting using an anti-p23, anti-p24, or anti-ERGIC53 antibody. VFG-GPI was detected with an anti-GFP antibody. Total lysate corresponding to 1% and immunoprecipitates were used for analysis.
Figure 5.
p24 family is required for the GPI-AP transport from the ER. (A) Relative amount of p23 and p24 mRNA from FF8 cells stably transfected with empty vector (+Vec) or p23 siRNA vector (+242) as determined by quantitative RT-PCR analysis. (B) Western blotting of p23 and p24 in p23 knockdown cells. FF8 cells permanently transfected with empty vector (Vec) or p23 siRNA vector (242) were lysed and the proteins were resolved by SDS-PAGE, followed by immunoblotting using rabbit anti-p23, anti-p24, or anti-ERGIC53 polyclonal antibody. (C) The cell surface expression of VFG-GPI was traced by flow cytometry. FF8 cells stably transfected with empty vector (+Vec) or p23 siRNA vector (+242) were stained with anti-Flag antibody at the indicated times after the commencement of transport. The geometric mean fluorescent values of surface VFG-GPI were quantified at each time point. The geometric mean of FF8 + Vec at 60 min was plotted as 100% relative transport. Values represent the mean ± SD (error bars; n = 3). (D and E) Pulse-chase transport analysis of Venus-CD59. Cells stably expressing Venus-CD59 were transfected with siRNAs against p23 or control RNA. After 72 h, the cells were pulse-labeled with [35S]methionine/cysteine followed by chasing for the indicated times, and immunoprecipitated with an antibody against Venus. Immunoprecipitates were treated with Endo-H, separated by SDS-PAGE, and analyzed and quantified using a Cyclone Phosphor Imager (Packard). The proportion of Golgi form (Endo-H–resistant form) and Venus-CD59 were plotted in E.
Figure 6.
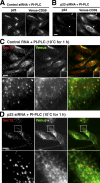
Sorting of GPI-APs to the ERES is dependent on p24 family. (A and B) CHO-K1 cells stably expressing Venus-CD59 were transfected with control siRNA (A) or siRNA against p23 (B). After 72 h, the cells were treated with PI-PLC, followed by fixation, permeabilization, and immunostaining with anti-p23. Arrows indicate efficiently silenced cells. (C and D) CHO-K1 cells stably expressing Venus-CD59 were transfected with control RNA (C) or siRNA against p23 (D). After 72 h, the cells were treated with PI-PLC, incubated at 10°C for 1 h, fixed with 4% paraformaldehyde, and stained with anti-Sec13 to observe the ERES. In the merged images, Sec13 and Venus-CD59 were shown in red and green, respectively. Enlarged views of the boxed regions are shown below. Bars, 10 µm.
Figure 7.
p24 proteins associated with GPI-APs in a pH-dependent manner. (A) Immunoprecipitation of VFG-GPI with p23 and p24 at various pH levels. FF8 cells were cultured with doxycycline at 40°C for 24 h to induce VFG-GPI expression and its accumulation in the ER. The cells were then incubated at 32°C for 20 min to initiate VFG-GPI transport. After cell lysis in lysis-IP buffer III of the indicated pH, VFG-GPI was precipitated with anti-Flag beads and washed five times in wash buffer III of the indicated pH, followed by immunoblotting using an anti-p23, anti-p24, or anti-ERGIC53 antibody. VFG-GPI was detected with an anti-GFP antibody. Total lysate corresponding to 1% and immunoprecipitates were used for analysis. Bands in 1% total fractions gradually increased with an increased pH partly because solubilization of proteins in 1% digitonin was slightly better at higher pH. (B) Release of p23 and p24 from VFG-GPI by lowering pH. FF8 cells prepared in a similar manner to A were lysed in lysis-IP buffer III, pH 7.4, and VFG-GPI was precipitated with anti-Flag beads. After washing five times with wash buffer III, pH 7.4, wash buffer III at the indicated pH was added and incubated at 4°C for 15 min. The supernatant (S) and bead pellets (P) were collected, followed by immunoblotting as in A.
Figure 8.
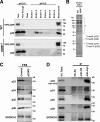
p24 proteins form heteromeric complexes with subfamily members. (A and B) Identification of proteins associated with VFG-GPI depending on pH. FF8 (WT) and FPRC2 (pgap1) cells were cultured with doxycycline at 40°C for 24 h. The cells were then incubated at 32°C for 20 min. After cell lysis using lysis-IP buffer II, pH 8.0, VFG-GPI was purified using an anti-Flag column. After thorough washing in wash buffer II, pH 8.0, the binding proteins were eluted with elution buffer II, pH 6.0. After elution with six bed volumes of buffer, proteins bound to the column were extracted with SDS sample buffer. Each fraction was subjected to SDS-PAGE, followed by immunoblotting using an anti-GFP or anti-p23 antibody (A). Proteins in eluted fraction 2 from FF8 (WT) and FPRC2 (pgap1) were detected by silver staining (B). Protein bands at 20 and 25 kD were identified by mass spectrometry as Tmed10 (p23), Tmed2 (p24), Tmed9 (p25), and Tmed5 (p28). Detected fragments are shown in Fig. S4. Protein bands indicated by an asterisk (*) were observed through all fractions (elutions 1−6) at similar levels and were not specific in WT cells. (C) Knockdown of p23 destabilized other p24 proteins. FF8 cells permanently transfected with an empty vector (Control) or p23 siRNA vector (p23KD) were lysed, and proteins were resolved by SDS-PAGE, followed by immunoblotting using rabbit anti-p23, anti-p24, anti-p25, anti-p28, and anti-ERGIC53 polyclonal antibodies. (D) Coimmunoprecipitation of myc-p23 with p24 proteins. FF8 cells were stably transfected with a retrovirus vector expressing RNAi-resistant myc-tagged p23 (myc-p23) and shRNA against endogenous p23, as described in
Fig. S5
. After cell lysis, myc-p23 was precipitated with anti-HA (control) or anti-myc antibodies or without antibody (No Ab), and coprecipitated proteins were detected by immunoblotting against anti-p23, anti-p24, anti-p25, anti-p28, and anti-ERGIC53 antibodies. Total lysate corresponding to 4% and immunoprecipitates were used for analysis. *, IgG heavy chains.
Figure 9.
A model of selective sorting and transport of GPI-APs from the ER. After GPI transfer to proteins by the GPI transamidase, the acyl chain linked to inositol in the GPI anchor is eliminated by PGAP1 (1), and a side-chain EtNP on the second mannose of the GPI anchor is removed by PGAP5 (2). These two GPI remodeling reactions in the ER are critical for the sorting of GPI-APs to the ERES. The remodeled GPI-APs are efficiently recognized by the p24 protein family complex that concentrates GPI-APs into the COPII-derived vesicles (3). After transport to the ERGIC or the cis-Golgi, GPI-APs dissociate from the p24 protein family complex because of decreased luminal pH in these compartments (4). The p24 complexes are retrieved from the Golgi to the ER by the COPI vesicles (5).
Similar articles
- The yeast p24 complex regulates GPI-anchored protein transport and quality control by monitoring anchor remodeling.
Castillon GA, Aguilera-Romero A, Manzano-Lopez J, Epstein S, Kajiwara K, Funato K, Watanabe R, Riezman H, Muñiz M. Castillon GA, et al. Mol Biol Cell. 2011 Aug 15;22(16):2924-36. doi: 10.1091/mbc.E11-04-0294. Epub 2011 Jun 16. Mol Biol Cell. 2011. PMID: 21680708 Free PMC article. - COPII coat composition is actively regulated by luminal cargo maturation.
Manzano-Lopez J, Perez-Linero AM, Aguilera-Romero A, Martin ME, Okano T, Silva DV, Seeberger PH, Riezman H, Funato K, Goder V, Wellinger RE, Muñiz M. Manzano-Lopez J, et al. Curr Biol. 2015 Jan 19;25(2):152-162. doi: 10.1016/j.cub.2014.11.039. Epub 2014 Dec 31. Curr Biol. 2015. PMID: 25557665 - Transport of glycosylphosphatidylinositol-anchored proteins from the endoplasmic reticulum.
Kinoshita T, Maeda Y, Fujita M. Kinoshita T, et al. Biochim Biophys Acta. 2013 Nov;1833(11):2473-8. doi: 10.1016/j.bbamcr.2013.01.027. Epub 2013 Feb 1. Biochim Biophys Acta. 2013. PMID: 23380706 Review. - Exit of GPI-anchored proteins from the ER differs in yeast and mammalian cells.
Rivier AS, Castillon GA, Michon L, Fukasawa M, Romanova-Michaelides M, Jaensch N, Hanada K, Watanabe R. Rivier AS, et al. Traffic. 2010 Aug;11(8):1017-33. doi: 10.1111/j.1600-0854.2010.01081.x. Epub 2010 May 11. Traffic. 2010. PMID: 20477992 - Endoplasmic Reticulum Export of GPI-Anchored Proteins.
Lopez S, Rodriguez-Gallardo S, Sabido-Bozo S, Muñiz M. Lopez S, et al. Int J Mol Sci. 2019 Jul 17;20(14):3506. doi: 10.3390/ijms20143506. Int J Mol Sci. 2019. PMID: 31319476 Free PMC article. Review.
Cited by
- The many hats of transmembrane emp24 domain protein TMED9 in secretory pathway homeostasis.
Roberts BS, Satpute-Krishnan P. Roberts BS, et al. Front Cell Dev Biol. 2023 Jan 16;10:1096899. doi: 10.3389/fcell.2022.1096899. eCollection 2022. Front Cell Dev Biol. 2023. PMID: 36733337 Free PMC article. Review. - Protein lipidation in health and disease: molecular basis, physiological function and pathological implication.
Yuan Y, Li P, Li J, Zhao Q, Chang Y, He X. Yuan Y, et al. Signal Transduct Target Ther. 2024 Mar 15;9(1):60. doi: 10.1038/s41392-024-01759-7. Signal Transduct Target Ther. 2024. PMID: 38485938 Free PMC article. Review. - ER cargo properties specify a requirement for COPII coat rigidity mediated by Sec13p.
Copic A, Latham CF, Horlbeck MA, D'Arcangelo JG, Miller EA. Copic A, et al. Science. 2012 Mar 16;335(6074):1359-62. doi: 10.1126/science.1215909. Epub 2012 Feb 2. Science. 2012. PMID: 22300850 Free PMC article. - The yeast p24 complex regulates GPI-anchored protein transport and quality control by monitoring anchor remodeling.
Castillon GA, Aguilera-Romero A, Manzano-Lopez J, Epstein S, Kajiwara K, Funato K, Watanabe R, Riezman H, Muñiz M. Castillon GA, et al. Mol Biol Cell. 2011 Aug 15;22(16):2924-36. doi: 10.1091/mbc.E11-04-0294. Epub 2011 Jun 16. Mol Biol Cell. 2011. PMID: 21680708 Free PMC article. - Mitochondrial Dysfunction Plus High-Sugar Diet Provokes a Metabolic Crisis That Inhibits Growth.
Kemppainen E, George J, Garipler G, Tuomela T, Kiviranta E, Soga T, Dunn CD, Jacobs HT. Kemppainen E, et al. PLoS One. 2016 Jan 26;11(1):e0145836. doi: 10.1371/journal.pone.0145836. eCollection 2016. PLoS One. 2016. PMID: 26812173 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous