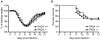PAD4-mediated neutrophil extracellular trap formation is not required for immunity against influenza infection - PubMed (original) (raw)
PAD4-mediated neutrophil extracellular trap formation is not required for immunity against influenza infection
Saskia Hemmers et al. PLoS One. 2011.
Abstract
During an inflammatory response, neutrophils migrate to the site of infection where they can kill invading pathogens by phagocytosis, secretion of anti-microbicidal mediators or the release of neutrophil extracellular traps (NETs). NETs are specialized anti-microbial structures comprised of decondensed chromatin decorated with microbicidal agents. Increased amount of NETs have been found in patients suffering from the chronic lung inflammatory disease cystic fibrosis, correlating with increased severity of pulmonary obstruction. Furthermore, acute lung inflammation during influenza A infection is characterized by a massive influx of neutrophils into the lung. The role of NETs during virus-mediated lung inflammation is unknown. Peptidylarginine deiminase 4 (PAD4)-mediated deimination of histone H3 and H4 is required for NET formation. Therefore, we generated a PAD4-deficient mouse strain that has a striking inability to form NETs. These mice were infected with influenza A/WSN, and the disease was monitored at the level of leukocytic lung infiltration, lung pathology, viral replication, weight loss and mortality. PAD4 KO fared comparable to WT mice in all the parameters tested, but they displayed slight but statistically different weight loss kinetics during infection that was not reflected in enhanced survival. Overall, we conclude that PAD4-mediated NET formation is dispensable in a mouse model of influenza A infection.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Figure 1. Generation and characterization of the PAD4 conditional knockout mouse strain.
(A) Schematic depiction of the targeted PAD4 genomic locus. Exons are depicted as numbered boxes. (B) Genomic DNA from three ES cell clones was digested with _Hind_III and analyzed by Southern blot with a labeled 5′ probe to confirm targeting of the PAD4 locus. (C) Mouse genomic tail DNA was screened for integration of the targeting vector into the PAD4 locus by Southern blotting. Genotypes as identified by PCR are indicated on top. (D) Cells from the peritoneal cavity of PAD4 WT or PAD4 KO mice were isolated and analyzed by immunoblotting for the presence of PAD4. The membrane was reprobed with anti-tubulin to ensure equal loading between samples.
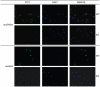
Figure 2. PAD4 is required for histone deimination.
Casein-elicited neutrophils from PAD4 WT and KO mice were analyzed by immunofluorescence for the presence of PAD4 (α huPAD4) or deiminated histone H4 (α H4R3cit) in the FITC channel (left panel) and counterstained with DAPI to visualize DNA content (middle panel). Representative pictures from two independent experiments are shown. All pictures were taken with the 40× objective.
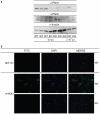
Figure 3. PAD4 expression and activity is detectable in the lung after influenza infection.
PAD4 WT and KO mice were infected with 2000 PFU influenza A/WSN intra-tracheally (i.t.). Lungs were harvested at d3 and d7 post infection (p.i.) (5 mice/group at each timepoint) and leukocytes were isolated as described in Materials and Methods. (A) Leukocytes from individual (d3 p.i.) or pooled mice (d7 p.i.) were lysed and analyzed for PAD2 or PAD4 expression by immunoblotting. Protein loading was assessed by comparing β-actin levels in the lysates. (B) Lung leukocytes from d3 p.i. were adhered to coverslips and analyzed for deiminated histone H4 levels (α H4cit3) by immunofluorescence. Cells were counterstained with DAPI to visualize DNA. Pictures were taken with the 20× objective.
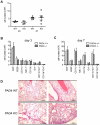
Figure 4. Lung leukocyte infiltration during influenza infection is comparable between PAD4 WT and KO mice.
Experimental setup as described in Figure 2. (A–C) Five mice per group were analyzed at d3 or d7 post infection. Leukocytes were isolated from infected lungs. (A) Total cell numbers of lung-infiltrating leukocytes at d3 and d7 p.i. Each symbol represents an individual mouse, filled squares represent PAD4 WT, open squares depict PAD4 KO mice. (B+C) The subsets of infiltrating leukocytes were enumerated by flow cytometry both at d3 p.i. (B) and d7 p.i. (C). Gated populations are indicated at the bottom of the graph. Total numbers of infiltrating cells are shown. WT mice are depicted in white bars, KO mice as grey bars. Bars represent mean + SEM. (D) Lungs for histological examination were harvested at d8 p.i. (5 mice/group). Leukocyte infiltration was assessed on H&E stained paraffin sections. Sections from two representative mice per group are shown. The scale bar indicates 200 µm. Data is representative of two independent experiments.
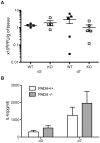
Figure 5. Viral titers and proinflammatory cytokine levels are similar between wildtype and PAD4-deficient mice.
Experimental setup as decribed in Figure 2. Lung homogenates from five mice per group were analyzed at d3 or d7 post infection with influenza A/WSN. (A) Viral titers were assessed by a plaque-forming assay on MDCK cells. Symbols represent individual animals (filled squares = PAD4 WT; open squares = PAD4 KO). Means +/− SEM are indicated. (B) Interleukin-6 levels were analyzed by tissue homogenate ELISA. Data is adjusted to starting tissue weight. Bars represent a group of eight mice as mean + SEM (white bars = PAD4 WT; grey bars = PAD4 KO). Data are representative of two individual experiments.
Figure 6. PAD4-deficient mice display a slight decrease in weight loss upon influenza infection but similar survival kinetics.
Mice were infected with 2000 PFU influenza A/WSN i.t. and monitored daily for weight loss and morbidity. Data represented here includes 19 WT mice and 19 PAD4 KO mice and is the result of combining two individual experiments. (A) Mice were weighed daily and weight loss is depicted as percent of starting weight. Timepoints are plotted as mean +/− SEM, filled squares represent WT mice and open squares depict PAD KO mice. Significant differences between the groups are indictated by asteriks (p<0.05 = *; p<0.01 = **; p<0.001 = ***) (B) Survival was monitored daily for 21d p.i. The data is plotted as percent survival over time and statistical analysis was performed using the log-rank test (p = 0.9983). The dashed line indicates 50% survival. Filled squares represent WT mice, open squares depict PAD4 KO mice.
Similar articles
- PAD4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps.
Li P, Li M, Lindberg MR, Kennett MJ, Xiong N, Wang Y. Li P, et al. J Exp Med. 2010 Aug 30;207(9):1853-62. doi: 10.1084/jem.20100239. Epub 2010 Aug 23. J Exp Med. 2010. PMID: 20733033 Free PMC article. - PAD4 Deficiency Improves Bleomycin-induced Neutrophil Extracellular Traps and Fibrosis in Mouse Lung.
Suzuki M, Ikari J, Anazawa R, Tanaka N, Katsumata Y, Shimada A, Suzuki E, Tatsumi K. Suzuki M, et al. Am J Respir Cell Mol Biol. 2020 Dec;63(6):806-818. doi: 10.1165/rcmb.2019-0433OC. Am J Respir Cell Mol Biol. 2020. PMID: 32915635 - PAD4-deficiency does not affect bacteremia in polymicrobial sepsis and ameliorates endotoxemic shock.
Martinod K, Fuchs TA, Zitomersky NL, Wong SL, Demers M, Gallant M, Wang Y, Wagner DD. Martinod K, et al. Blood. 2015 Mar 19;125(12):1948-56. doi: 10.1182/blood-2014-07-587709. Epub 2015 Jan 26. Blood. 2015. PMID: 25624317 Free PMC article. - Neutrophil Extracellular Traps in Pulmonary Diseases: Too Much of a Good Thing?
Porto BN, Stein RT. Porto BN, et al. Front Immunol. 2016 Aug 15;7:311. doi: 10.3389/fimmu.2016.00311. eCollection 2016. Front Immunol. 2016. PMID: 27574522 Free PMC article. Review. - Progression of Cystic Fibrosis Lung Disease from Childhood to Adulthood: Neutrophils, Neutrophil Extracellular Trap (NET) Formation, and NET Degradation.
Khan MA, Ali ZS, Sweezey N, Grasemann H, Palaniyar N. Khan MA, et al. Genes (Basel). 2019 Feb 26;10(3):183. doi: 10.3390/genes10030183. Genes (Basel). 2019. PMID: 30813645 Free PMC article. Review.
Cited by
- A Hairy Cituation - PADIs in Regeneration and Alopecia.
Vikhe Patil K, Mak KH, Genander M. Vikhe Patil K, et al. Front Cell Dev Biol. 2021 Dec 13;9:789676. doi: 10.3389/fcell.2021.789676. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34966743 Free PMC article. Review. - In vivo evidence for extracellular DNA trap formation.
Yousefi S, Simon D, Stojkov D, Karsonova A, Karaulov A, Simon HU. Yousefi S, et al. Cell Death Dis. 2020 Apr 30;11(4):300. doi: 10.1038/s41419-020-2497-x. Cell Death Dis. 2020. PMID: 32355207 Free PMC article. Review. - Neutrophil extracellular trap inhibition increases inflammation, bacteraemia and mortality in murine necrotizing enterocolitis.
Chaaban H, Burge K, Eckert J, Keshari RS, Silasi R, Lupu C, Warner B, Escobedo M, Caplan M, Lupu F. Chaaban H, et al. J Cell Mol Med. 2021 Dec;25(23):10814-10824. doi: 10.1111/jcmm.15338. Epub 2020 Jun 8. J Cell Mol Med. 2021. PMID: 32515131 Free PMC article. - Neutrophil Extracellular Traps in Coronavirus Disease-19-Associated Ischemic Stroke: A Novel Avenue in Neuroscience.
Pramitasuri TI, Laksmidewi AAAP, Putra IBK, Dalimartha FA. Pramitasuri TI, et al. Exp Neurobiol. 2021 Feb 28;30(1):1-12. doi: 10.5607/en20048. Exp Neurobiol. 2021. PMID: 33632982 Free PMC article. - Caught in a Trap? Proteomic Analysis of Neutrophil Extracellular Traps in Rheumatoid Arthritis and Systemic Lupus Erythematosus.
Chapman EA, Lyon M, Simpson D, Mason D, Beynon RJ, Moots RJ, Wright HL. Chapman EA, et al. Front Immunol. 2019 Mar 11;10:423. doi: 10.3389/fimmu.2019.00423. eCollection 2019. Front Immunol. 2019. PMID: 30915077 Free PMC article.
References
- Nathan C. Neutrophils and immunity: challenges and opportunities. Nature reviews Immunology. 2006;6:173–182. - PubMed
- Borregaard N. Neutrophils, from marrow to microbes. Immunity. 2010;33:657–670. - PubMed
- Nauseef WM. How human neutrophils kill and degrade microbes: an integrated view. Immunol Rev. 2007;219:88–102. - PubMed
- Flannagan RS, Cosio G, Grinstein S. Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nature reviews Microbiology. 2009;7:355–366. - PubMed
- Scapini P, Lapinet-Vera JA, Gasperini S, Calzetti F, Bazzoni F, et al. The neutrophil as a cellular source of chemokines. Immunol Rev. 2000;177:195–203. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 AI007354/AI/NIAID NIH HHS/United States
- AI74564/AI/NIAID NIH HHS/United States
- AI067460/AI/NIAID NIH HHS/United States
- AI007354/AI/NIAID NIH HHS/United States
- AI09484/AI/NIAID NIH HHS/United States
- R01 AI009484/AI/NIAID NIH HHS/United States
- R01 AI067460/AI/NIAID NIH HHS/United States
- U01 AI074564/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials