Emerging roles of TET proteins and 5-hydroxymethylcytosines in active DNA demethylation and beyond - PubMed (original) (raw)
Review
Emerging roles of TET proteins and 5-hydroxymethylcytosines in active DNA demethylation and beyond
Junjie U Guo et al. Cell Cycle. 2011.
Abstract
Cytosine methylation is the major epigenetic modification of metazoan DNA. Although there is strong evidence that active DNA demethylation occurs in animal cells, the molecular details of this process are unknown. The recent discovery of the TET protein family (TET1-3) 5-methylcytosine hydroxylases has provided a new entry point to reveal the identity of the long-sought DNA demethylase. Here, we review the recent progress in understanding the function of TET proteins and 5-hydroxymethylcytosine (5hmC) through various biochemical and genomic approaches, the current evidence for a role of 5hmC as an early intermediate in active DNA demethylation and the potential functions of TET proteins and 5hmC beyond active DNA demethylation. We also discuss how future studies can extend our knowledge of this novel epigenetic modification.
Figures
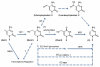
Figure 1
Potential mechanisms for active DNA demethylation through 5hmC. (1) Evidence suggests that 5hmC may be converted to 5hmU by AID/APOBEC deaminases and repaired by the base excision repair (BER) pathway. (2) 5hmC may also be further oxidized to 5-carboxylcytosine and decarboxylated/repaired to C. (3) A 5hmC glycosylases may excise 5hmC and directly initiate BER. (4) Direct conversion from 5hmC to C may also occur. (5) Nuclear excision repair (NER) pathway may excise 5hmC-containing DNA strands. Besides promoting DNA demethylation, 5hmC may regulate transcription through unidentified 5hmC-binding proteins. Solid arrows indicate catalytic steps that are supported by experimental evidence, whereas dashed arrows indicate speculative processes.
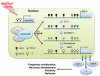
Figure 2
Neuronal activity-induced DNA demethylation in the adult brain. Neuronal activation in the dentate gyrus induces expression of Gadd45b, which plays an essential role in demethylation of Bdnf and Fgf1 promoters. This process also involves Tet1 and Apobec1. Gadd45b-dependent DNA demethylation leads to expression of important genes, such as Bdnf and Fgf1, and can potentially have a broad impact in the mature nervous system.
Similar articles
- GADD45a physically and functionally interacts with TET1.
Kienhöfer S, Musheev MU, Stapf U, Helm M, Schomacher L, Niehrs C, Schäfer A. Kienhöfer S, et al. Differentiation. 2015 Jul-Oct;90(1-3):59-68. doi: 10.1016/j.diff.2015.10.003. Epub 2015 Nov 3. Differentiation. 2015. PMID: 26546041 Free PMC article. - Role of ten-eleven translocation proteins and 5-hydroxymethylcytosine in hepatocellular carcinoma.
Wang P, Yan Y, Yu W, Zhang H. Wang P, et al. Cell Prolif. 2019 Jul;52(4):e12626. doi: 10.1111/cpr.12626. Epub 2019 Apr 29. Cell Prolif. 2019. PMID: 31033072 Free PMC article. Review. - Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells.
Blaschke K, Ebata KT, Karimi MM, Zepeda-Martínez JA, Goyal P, Mahapatra S, Tam A, Laird DJ, Hirst M, Rao A, Lorincz MC, Ramalho-Santos M. Blaschke K, et al. Nature. 2013 Aug 8;500(7461):222-6. doi: 10.1038/nature12362. Epub 2013 Jun 30. Nature. 2013. PMID: 23812591 Free PMC article. - Tet family proteins and 5-hydroxymethylcytosine in development and disease.
Tan L, Shi YG. Tan L, et al. Development. 2012 Jun;139(11):1895-902. doi: 10.1242/dev.070771. Development. 2012. PMID: 22569552 Free PMC article. Review. - Structural insight into substrate preference for TET-mediated oxidation.
Hu L, Lu J, Cheng J, Rao Q, Li Z, Hou H, Lou Z, Zhang L, Li W, Gong W, Liu M, Sun C, Yin X, Li J, Tan X, Wang P, Wang Y, Fang D, Cui Q, Yang P, He C, Jiang H, Luo C, Xu Y. Hu L, et al. Nature. 2015 Nov 5;527(7576):118-22. doi: 10.1038/nature15713. Epub 2015 Oct 28. Nature. 2015. PMID: 26524525
Cited by
- TET1 is controlled by pluripotency-associated factors in ESCs and downmodulated by PRC2 in differentiated cells and tissues.
Neri F, Incarnato D, Krepelova A, Dettori D, Rapelli S, Maldotti M, Parlato C, Anselmi F, Galvagni F, Oliviero S. Neri F, et al. Nucleic Acids Res. 2015 Aug 18;43(14):6814-26. doi: 10.1093/nar/gkv392. Epub 2015 Apr 29. Nucleic Acids Res. 2015. PMID: 25925565 Free PMC article. - 5-Hydroxymethylcytosine in cord blood and associations of DNA methylation with sex in newborns.
Solomon O, Macisaac JL, Tindula G, Kobor MS, Eskenazi B, Holland N. Solomon O, et al. Mutagenesis. 2019 Dec 19;34(4):315-322. doi: 10.1093/mutage/gez023. Mutagenesis. 2019. PMID: 31587037 Free PMC article. - Brain Functional Reserve in the Context of Neuroplasticity after Stroke.
Dąbrowski J, Czajka A, Zielińska-Turek J, Jaroszyński J, Furtak-Niczyporuk M, Mela A, Poniatowski ŁA, Drop B, Dorobek M, Barcikowska-Kotowicz M, Ziemba A. Dąbrowski J, et al. Neural Plast. 2019 Feb 27;2019:9708905. doi: 10.1155/2019/9708905. eCollection 2019. Neural Plast. 2019. PMID: 30936915 Free PMC article. Review. - DNA-methylation gene network dysregulation in peripheral blood lymphocytes of schizophrenia patients.
Auta J, Smith RC, Dong E, Tueting P, Sershen H, Boules S, Lajtha A, Davis J, Guidotti A. Auta J, et al. Schizophr Res. 2013 Oct;150(1):312-8. doi: 10.1016/j.schres.2013.07.030. Epub 2013 Aug 12. Schizophr Res. 2013. PMID: 23938174 Free PMC article. - Deletion of Tet proteins results in quantitative disparities during ESC differentiation partially attributable to alterations in gene expression.
Reimer M Jr, Pulakanti K, Shi L, Abel A, Liang M, Malarkannan S, Rao S. Reimer M Jr, et al. BMC Dev Biol. 2019 Jul 8;19(1):16. doi: 10.1186/s12861-019-0196-6. BMC Dev Biol. 2019. PMID: 31286885 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 HD069184/HD/NICHD NIH HHS/United States
- R01 AG024984/AG/NIA NIH HHS/United States
- AG024984/AG/NIA NIH HHS/United States
- NS048271/NS/NINDS NIH HHS/United States
- R37 NS047344/NS/NINDS NIH HHS/United States
- R56 NS047344/NS/NINDS NIH HHS/United States
- R01 NS048271/NS/NINDS NIH HHS/United States
- R01 NS047344/NS/NINDS NIH HHS/United States
- HD069184/HD/NICHD NIH HHS/United States
- NS047344/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources