Oncogenic K-Ras decouples glucose and glutamine metabolism to support cancer cell growth - PubMed (original) (raw)
Oncogenic K-Ras decouples glucose and glutamine metabolism to support cancer cell growth
Daniela Gaglio et al. Mol Syst Biol. 2011.
Abstract
Oncogenes such as K-ras mediate cellular and metabolic transformation during tumorigenesis. To analyze K-Ras-dependent metabolic alterations, we employed ¹³C metabolic flux analysis (MFA), non-targeted tracer fate detection (NTFD) of ¹⁵N-labeled glutamine, and transcriptomic profiling in mouse fibroblast and human carcinoma cell lines. Stable isotope-labeled glucose and glutamine tracers and computational determination of intracellular fluxes indicated that cells expressing oncogenic K-Ras exhibited enhanced glycolytic activity, decreased oxidative flux through the tricarboxylic acid (TCA) cycle, and increased utilization of glutamine for anabolic synthesis. Surprisingly, a non-canonical labeling of TCA cycle-associated metabolites was detected in both transformed cell lines. Transcriptional profiling detected elevated expression of several genes associated with glycolysis, glutamine metabolism, and nucleotide biosynthesis upon transformation with oncogenic K-Ras. Chemical perturbation of enzymes along these pathways further supports the decoupling of glycolysis and TCA metabolism, with glutamine supplying increased carbon to drive the TCA cycle. These results provide evidence for a role of oncogenic K-Ras in the metabolic reprogramming of cancer cells.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Figure 1
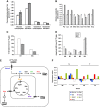
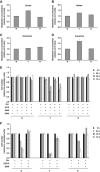
K-Ras transformed fibroblasts decouple glycolysis and the TCA cycle. (A) Extracellular uptake and secretion of Glc, Lac, Gln, and Glu in N and T cells for 54 h of growth. (B) Relative metabolite abundances in N and T cells measured by GC/MS. (C) M2 labeling of Cit and Glu in N and T cells cultured with a 1:1 mixture of [1-13C]glucose and [U-13C6]glucose. (D) Mass isotopomer distribution (MID) of Asp in cells cultured in the presence of [U-13C5]glutamine. Error bars indicate s.e.m. (_n_=3). (E) Schematic representation of the metabolic routes involved in M3-M4 Asp labeling by using [U13C5]glutamine tracer. In the scheme are represented the key cytoplasmic and mitochondrial enzymes involved in these metabolic routes. (F) Absolute expression values of some genes, involved in the pathways described in the scheme (E), in N and T cell lines. The two cell lines, grown in 25 mM Glc+4 mM Gln (normal medium), were collected for transcriptional analysis at two different time points (48 and 72 h). The list of gene abbreviations is available in Supplementary information.
Figure 2
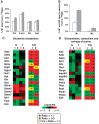
Labeled metabolites identified by NTFD analysis in cells cultured with [α-15N]glutamine for 54 h. (A) M1 labeling on amino acids and derivative metabolites in N (□) and T (▪) cells. (B) M1 and M2 labeling observed in free adenine in N (□) and T (▪) cells. (C) Heat maps of genes involved in Gln metabolism of N (blue line) and T (red line) cell lines, and corresponding ratios (T/N) between the two cell lines grown in normal medium and collected at two different time points (1=48 h and 2=72 h). (D) Heat maps of genes involved in purine metabolism of N (blue line) and T (red line) cell lines, and corresponding ratios (T/N) between the two cell lines grown in normal medium and collected at two different time points (1=48 h and 2=72 h). Color scale bar indicates the normalized expression intensity (log scale); in particular, low expression in green, high expression in red, and not changed expression in black. The ratio values are represented as Up—red color—when the ratio is ⩾1.1, down—green color—when the ratio is ⩽0.9 and no change—yellow color—when the ratio is between 0.9 and 1.1. The list of gene abbreviations is available in Supplementary information.
Figure 3
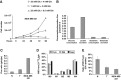
MDA-MB-231 human cancer cell line exhibits enhanced glycolysis and reduced TCA cycle activity. (A) MDA-MB-231 cells were plated at 3000 cells/cm2 in 6-well plates in normal medium. Culture medium was replaced after 18 h with normal medium (◊), or a medium containing 1 mM Glc+4 mM Gln (□) or 25 mM Glc+0.5 mM Gln (▵). Then, the cells were collected and counted at indicate time points. (B) Extracellular uptake of Glc and Gln and secretion of Lac and Glu after 54 h. (C) Ratio of Glc uptake to Gln uptake. (D) MID of Fum, Mal, and Asp in cells cultured in the presence of [U-13C5]glutamine. (E) Ratio of PDH flux to Glc uptake. Error bars indicate s.e.m. (_n_=3) using propagation of error.
Figure 4
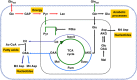
Schematic integrative representation of metabolic fluxes and transcriptional data in normal and mouse K-Ras transformed cells. (A, B) Integration maps of metabolic routes identified in T cells as compared with N cells by using flux analysis and transcriptional data. The ratio between T and N cells of both metabolic fluxes and gene expression values are represented by a color code (as indicated in the upper right legends) as follow: black (no change value), red (increased value), and green (decreased value) for flux analysis and Up—red color—T/N ratio ⩾1.1, down—green color—T/N ratio ⩽0.9 and no change—yellow color—T/N ratio between 0.9 and 1.1 for transcriptional data. The list of gene abbreviations used in the figure can be found in ‘Supplementary information’. Metabolic flux analysis of central carbon metabolism was obtained by using the data derived from [U-13C5] glutamine, a mixture of [U-13C6] glucose and 1-13C] glucose (A) and [α-15N]glutamine (B) as specific isotopic tracers. The net and exchange fluxes were calculated as described under ‘Materials and methods’. Transcriptional data of N and T regulated genes were obtained as described in ‘Materials and methods’.
Figure 5
Relative metabolites concentrations in N, T, and R cell lines and effect of aminooxyacetate (AOA) and epigallocatechin gallate (EGCG) treatments on their proliferation. Evaluation of Cit (A), Mal (B), Glu (C), and Asp (D) intracellular concentrations was carried out after 54 h of grown in normal medium by using enzymatic assays. Analysis of AOA (E) and EGCG (F) treatments on N, T, and R cell lines proliferation. Cells, seeded at density of 3000 cells/cm2, after 18 h and following a complete medium replacement, were treated with 100 μM AOA, or 1 mM dimethyl aspartate (DMD), or 2 mM dimethyl α-ketoglutarate (DMK) or 20 μM EGCG and counted at indicated time points. Error bars indicate s.e.m. (_n_=3).
Figure 6
(A) Effects of EGCG inhibitor at short term in N, T, and R cell lines. Cells, seeded at density of 3000 cells/cm2, after 18 h and following a complete medium replacement, were treated with 20 μM EGCG and/or 5 mM NAC and counted at indicated time points. (B) Analysis of cell death by fluorescence microscopy using PI (red color), Annexin V (green color), and Hoechst for nuclei staining (Blue color). The cells were plated at 3000 cells/cm2 on microglasses, treated as above described and analyzed after 8 h. The images were acquired by × 60 objective using Metamorph 7 software. (C) Analysis of intracellular ATP level by enzymatic assays. The cells were treated as above described and collected at indicated time points. (D) Intracellular ROS levels were measured by using 5 mM DCFH2-DA staining. The cells were treated as above described and collected at indicated time points. Error bars indicate s.e.m. (_n_=3).
Figure 7
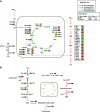
Schematic representation of Glc and Gln pathways decoupling in K-Ras cancer cells. Glc provides the main energy source for cancer cells with oncogenic K-Ras. Instead, Gln provides the carbon and nitrogen for the synthesis of biomass building blocks (amino acids, nucleotides, and fatty acids). Red arrows represent enhanced flux, green arrows represent reduced flux, black arrows represent no change flux, blue arrow represents forward flux of TCA cycle, and sky-blue represents reductive carboxylation flux.
Similar articles
- Altered glucose metabolism in Harvey-ras transformed MCF10A cells.
Zheng W, Tayyari F, Gowda GA, Raftery D, McLamore ES, Porterfield DM, Donkin SS, Bequette B, Teegarden D. Zheng W, et al. Mol Carcinog. 2015 Feb;54(2):111-20. doi: 10.1002/mc.22079. Epub 2013 Sep 2. Mol Carcinog. 2015. PMID: 24000146 Free PMC article. - Blocking anaplerotic entry of glutamine into the TCA cycle sensitizes K-Ras mutant cancer cells to cytotoxic drugs.
Saqcena M, Mukhopadhyay S, Hosny C, Alhamed A, Chatterjee A, Foster DA. Saqcena M, et al. Oncogene. 2015 May 14;34(20):2672-80. doi: 10.1038/onc.2014.207. Epub 2014 Jul 14. Oncogene. 2015. PMID: 25023699 Free PMC article. - Analysis of Melanoma Cell Glutamine Metabolism by Stable Isotope Tracing and Gas Chromatography-Mass Spectrometry.
Scott DA. Scott DA. Methods Mol Biol. 2021;2265:91-110. doi: 10.1007/978-1-0716-1205-7_7. Methods Mol Biol. 2021. PMID: 33704708 - The molecular determinants of de novo nucleotide biosynthesis in cancer cells.
Tong X, Zhao F, Thompson CB. Tong X, et al. Curr Opin Genet Dev. 2009 Feb;19(1):32-7. doi: 10.1016/j.gde.2009.01.002. Epub 2009 Feb 5. Curr Opin Genet Dev. 2009. PMID: 19201187 Free PMC article. Review. - Metabolic reprogramming in cancer: unraveling the role of glutamine in tumorigenesis.
Daye D, Wellen KE. Daye D, et al. Semin Cell Dev Biol. 2012 Jun;23(4):362-9. doi: 10.1016/j.semcdb.2012.02.002. Epub 2012 Feb 11. Semin Cell Dev Biol. 2012. PMID: 22349059 Review.
Cited by
- Lipid droplet storage promotes murine pancreatic tumor growth.
Grachan JJ, Kery M, Giaccia AJ, Denko NC, Papandreou I. Grachan JJ, et al. Oncol Rep. 2021 Apr;45(4):21. doi: 10.3892/or.2021.7972. Epub 2021 Mar 2. Oncol Rep. 2021. PMID: 33649859 Free PMC article. - Expanding the concepts and tools of metabolic engineering to elucidate cancer metabolism.
Keibler MA, Fendt SM, Stephanopoulos G. Keibler MA, et al. Biotechnol Prog. 2012 Nov-Dec;28(6):1409-18. doi: 10.1002/btpr.1629. Epub 2012 Oct 18. Biotechnol Prog. 2012. PMID: 22961737 Free PMC article. Review. - Suppression of the HBP Function Increases Pancreatic Cancer Cell Sensitivity to a Pan-RAS Inhibitor.
Ricciardiello F, Bergamaschi L, De Vitto H, Gang Y, Zhang T, Palorini R, Chiaradonna F. Ricciardiello F, et al. Cells. 2021 Feb 18;10(2):431. doi: 10.3390/cells10020431. Cells. 2021. PMID: 33670598 Free PMC article. - Preclinical Evaluation of 4-[18F]Fluoroglutamine PET to Assess ASCT2 Expression in Lung Cancer.
Hassanein M, Hight MR, Buck JR, Tantawy MN, Nickels ML, Hoeksema MD, Harris BK, Boyd K, Massion PP, Manning HC. Hassanein M, et al. Mol Imaging Biol. 2016 Feb;18(1):18-23. doi: 10.1007/s11307-015-0862-4. Mol Imaging Biol. 2016. PMID: 25971659 Free PMC article. - Evolutionary origins and innovations sculpting the mammalian PRPS enzyme complex.
Karki BR, Macmillan AC, Vicente-Muñoz S, Greis KD, Romick LE, Cunningham JT. Karki BR, et al. bioRxiv [Preprint]. 2024 Oct 1:2024.10.01.616059. doi: 10.1101/2024.10.01.616059. bioRxiv. 2024. PMID: 39411161 Free PMC article. Preprint.
References
- Antoniewicz MR, Kelleher JK, Stephanopoulos G (2006) Determination of confidence intervals of metabolic fluxes estimated from stable isotope measurements. Metab Eng 8: 324–337 - PubMed
- Babushok VI, Linstrom PJ, Reed JJ, Zenkevich IG, Brown RL, Mallard WG, Stein SE (2007) Development of a database of gas chromatographic retention properties of organic compounds. J Chromatogr 1157: 414–421 - PubMed
- Baggetto LG (1992) Deviant energetic metabolism of glycolytic cancer cells. Biochimie 74: 959–974 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous