O-GlcNAc signalling: implications for cancer cell biology - PubMed (original) (raw)
Review
O-GlcNAc signalling: implications for cancer cell biology
Chad Slawson et al. Nat Rev Cancer. 2011.
Abstract
O-GlcNAcylation is the covalent attachment of β-D-N-acetylglucosamine (GlcNAc) sugars to serine or threonine residues of nuclear and cytoplasmic proteins, and it is involved in extensive crosstalk with other post-translational modifications, such as phosphorylation. O-GlcNAcylation is becoming increasing realized as having important roles in cancer-relevant processes, such as cell signalling, transcription, cell division, metabolism and cytoskeletal regulation. However, currently little is known about the specific roles of aberrant O-GlcNAcylation in cancer. In this Opinion article, we summarize the current understanding of O-GlcNAcylation in cancer and its emerging functions in transcriptional regulation at the level of chromatin and transcription factors.
Figures
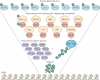
Figure 1. _O_-GlcNAc modifies the transcriptional machinery
In this inverted pyramid diagram, the transcription-regulating proteins that are modified by β-D-_N_-acetylglucosamine (GlcNAc; G) are shown. In addition to its catalytic role, _O_-linked _N_-acetylglucosamine transferase (OGT; red ovals) interacts with numerous transcription-regulating proteins. Starting from the bottom, histones are dynamically modified by _O_-GlcNAc; furthermore, the carboxy-terminal domain of RNA polymerase II (RNA Pol II), as well as the basal transcription factors (purple ovals) are also modified by _O_-GlcNAc. Many of the transcription-activating and transcription-repressing complexes (yellow circles) that contain histone-modifying enzymes, such as histone methyltransferases, histone acetytransferases and histone deacetylases, also interact with OGT, suggesting that OGT is an integral unit in the regulation of the histone code. Finally, many transcription factors (light blue circles) are modified by _O_-GlcNAc. Together, the collected data from many laboratories have strongly confirmed the regulation of gene transcription by _O_-GlcNAcylation. CEBPβ, CCAAT/enhancer-binding protein-β; CREB1, cyclicAMP-responsive element-binding protein 1; CRTC2, CREB-regulated transcription coactivator 2; FOXO, forkhead box protein O family; HCF1, host cell factor 1; MSL, male-specific lethal; NFAT, nuclear factor of activated T cells; NSL, nonspecific lethal; PDX1, pancreas/duodenum homeobox protein 1; PGC1α: peroxisome proliferator-activated receptor-γ co-activator 1α; PH, Polyhomeotic protein; TBP, TATA-box-binding protein.
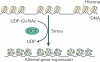
Figure 2. Histone _O_-GlcNAcylation increases after stress
After a cellular stress event, histone _O_-GlcNAcylation (the covalent attachment of β-D-_N_-acetylglucosamine (GlcNAc) sugars to serine or threonine residues of proteins; green dots) increases, and this is correlated with an increase in DNA compaction. Histone _O_-GlcNAcylation is catalysed by _O_-GlcNAc transferase (OGT) via the addition of a GlcNAc moiety from UDP-GlcNAc.
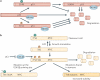
Figure 3. Regulation of transcription factors by _O_-GlcNAc
a | The tumour suppressor p53 associates with the protein MDM2, which keeps p53 at low levels in cells by promoting its degradation. Phosphorylation (P) at Thr155 stimulates the rapid degradation of p53, and _O_-GlcNAcylation (the covalent attachment of β-D-_N_-acetylglucosamine (GlcNAc) sugars to serine or threonine residues of nuclear and cytoplasmic proteins; G) at Ser149 disrupts MDM2 binding and promotes p53 stability. Mutations abolishing _O_-GlcNAc on p53 enhances degradation. b | In quiescent cells, the oncoprotein MYC is _O_-GlcNAcylated at Thr58. Upon growth signals, the _O_-GlcNAc modification is rapidly removed, and the protein is phosphorylated at Ser62. This site is a priming site for glycogen synthase kinase 3β (GSK3β), which can then phosphorylate Thr58, leading to the rapid degradation of MYC. Numerous solid tumours have mutations at Thr58, which promotes increased MYC stability and altered gene transcription. Inhibition of GSK3β by lithium chloride (LiCl) or mutations at Ser62 increases _O_-GlcNAcylation at Thr58.
Similar articles
- O-linked beta-N-acetylglucosamine (O-GlcNAc): Extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress.
Butkinaree C, Park K, Hart GW. Butkinaree C, et al. Biochim Biophys Acta. 2010 Feb;1800(2):96-106. doi: 10.1016/j.bbagen.2009.07.018. Epub 2009 Aug 6. Biochim Biophys Acta. 2010. PMID: 19647786 Free PMC article. Review. - O-GlcNAc signaling in cancer metabolism and epigenetics.
Singh JP, Zhang K, Wu J, Yang X. Singh JP, et al. Cancer Lett. 2015 Jan 28;356(2 Pt A):244-50. doi: 10.1016/j.canlet.2014.04.014. Epub 2014 Apr 24. Cancer Lett. 2015. PMID: 24769077 Free PMC article. Review. - Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease.
Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Hart GW, et al. Annu Rev Biochem. 2011;80:825-58. doi: 10.1146/annurev-biochem-060608-102511. Annu Rev Biochem. 2011. PMID: 21391816 Free PMC article. Review. - Emerging Role of Protein O-GlcNAcylation in Liver Metabolism: Implications for Diabetes and NAFLD.
Xie Z, Xie T, Liu J, Zhang Q, Xiao X. Xie Z, et al. Int J Mol Sci. 2023 Jan 21;24(3):2142. doi: 10.3390/ijms24032142. Int J Mol Sci. 2023. PMID: 36768465 Free PMC article. Review. - Implications and mechanisms of O-GlcNAcylation in cancer therapy resistance.
Gao H, Ma B, Zhao Y, Pan Y, Zhang A. Gao H, et al. Tumori. 2025 Feb;111(1):41-54. doi: 10.1177/03008916241299244. Epub 2024 Dec 24. Tumori. 2025. PMID: 39718082 Review.
Cited by
- O-GlcNAcylation of TDP-43 suppresses proteinopathies and promotes TDP-43's mRNA splicing activity.
Zhao MJ, Yao X, Wei P, Zhao C, Cheng M, Zhang D, Xue W, He WT, Xue W, Zuo X, Jiang LL, Luo Z, Song J, Shu WJ, Yuan HY, Liang Y, Sun H, Zhou Y, Zhou Y, Zheng L, Hu HY, Wang J, Du HN. Zhao MJ, et al. EMBO Rep. 2021 Jun 4;22(6):e51649. doi: 10.15252/embr.202051649. Epub 2021 Apr 15. EMBO Rep. 2021. PMID: 33855783 Free PMC article. - Hijacking the Hexosamine Biosynthetic Pathway to Promote EMT-Mediated Neoplastic Phenotypes.
Taparra K, Tran PT, Zachara NE. Taparra K, et al. Front Oncol. 2016 Apr 18;6:85. doi: 10.3389/fonc.2016.00085. eCollection 2016. Front Oncol. 2016. PMID: 27148477 Free PMC article. Review. - Discovery of O-GlcNAc-modified proteins in published large-scale proteome data.
Hahne H, Gholami AM, Kuster B. Hahne H, et al. Mol Cell Proteomics. 2012 Oct;11(10):843-50. doi: 10.1074/mcp.M112.019463. Epub 2012 Jun 1. Mol Cell Proteomics. 2012. PMID: 22661428 Free PMC article. - O-GlcNAcylation of MITF regulates its activity and CDK4/6 inhibitor resistance in breast cancer.
Zhang Y, Zhou S, Kai Y, Zhang YQ, Peng C, Li Z, Mughal MJ, Julie B, Zheng X, Ma J, Ma CX, Shen M, Hall MD, Li S, Zhu W. Zhang Y, et al. Nat Commun. 2024 Jul 3;15(1):5597. doi: 10.1038/s41467-024-49875-w. Nat Commun. 2024. PMID: 38961064 Free PMC article. - Photocrosslinking probes for capture of carbohydrate interactions.
Wu H, Kohler J. Wu H, et al. Curr Opin Chem Biol. 2019 Dec;53:173-182. doi: 10.1016/j.cbpa.2019.09.002. Epub 2019 Nov 6. Curr Opin Chem Biol. 2019. PMID: 31706134 Free PMC article. Review.
References
- Torres CR, Hart GW. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J. Biol. Chem. 1984;259:3308–3317. - PubMed
- Hart GW, Housley MP, Slawson C. Cycling of O-linked β-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–1022. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P20 RR024214/RR/NCRR NIH HHS/United States
- P20RR024214/RR/NCRR NIH HHS/United States
- R01 CA042486/CA/NCI NIH HHS/United States
- R37 HD013563/HD/NICHD NIH HHS/United States
- R24 DK084949/DK/NIDDK NIH HHS/United States
- R01 DK061671/DK/NIDDK NIH HHS/United States
- R01 DK61671/DK/NIDDK NIH HHS/United States
- P30 DK079637/DK/NIDDK NIH HHS/United States
- R01 CA42486/CA/NCI NIH HHS/United States
- N01‑HV‑00240/HV/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources