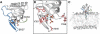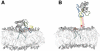Biophysical and computational studies of membrane penetration by the GRP1 pleckstrin homology domain - PubMed (original) (raw)
Biophysical and computational studies of membrane penetration by the GRP1 pleckstrin homology domain
Craig N Lumb et al. Structure. 2011.
Abstract
The pleckstrin homology (PH) domain of the general receptor for phosphoinositides 1 (GRP1) exhibits specific, high-affinity, reversible binding to phosphatidylinositol (3,4,5)-trisphosphate (PI(3,4,5)P(3)) at the plasma membrane, but the nature and extent of the interaction between this bound complex and the surrounding membrane environment remains unclear. Combining equilibrium and nonequilibrium molecular dynamics (MD) simulations, NMR spectroscopy, and monolayer penetration experiments, we characterize the membrane-associated state of GRP1-PH. MD simulations show loops flanking the binding site supplement the interaction with PI(3,4,5)P(3) through multiple contacts with the lipid bilayer. NMR data show large perturbations in chemical shift for these loop regions on binding to PI(3,4,5)P(3)-containing DPC micelles. Monolayer penetration experiments and further MD simulations demonstrate that mutating hydrophobic residues to polar residues in the flanking loops reduces membrane penetration. This supports a "dual-recognition" model of binding, with specific GRP1-PH-PI(3,4,5)P(3) interactions supplemented by interactions of loop regions with the lipid bilayer.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures
Graphical abstract
Figure 1
Structural Features of GRP1-PH (A) The three binding loops, β1/β2 (black), β3/β4 (green), and β6/β7 (blue), and their positions relative to the I(1,3,4,5)P4 head group. (B) The geometry of the I(1,3,4,5)P4 binding site, with the key amino acid residues referred to in the text shown as stick representations in pink. (C) The initial setup for the MD simulations, with the centers of the phosphate groups shown as gray spheres and the lipid bilayer depicted as a translucent white surface. Water molecules and ions are omitted for clarity. In all cases, the protein is shown as a ribbon diagram, and the I(1,3,4,5)P4 head group is shown as a stick model.
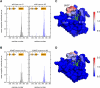
Figure 2
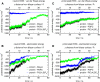
Membrane Penetration of GRP1-PH Observed during the MD Simulations (A) Mean number of nonpolar protein-lipid contacts per residue per ps over the 100 ns for the two wild-type protein simulation (one simulation in black and the second simulation in blue). (B) The same measure for the two A346E mutant simulations. (C) and (D) show these contacts projected on to the molecular surface of the protein for the wild-type and A346E mutants respectively, illustrating the decrease in penetration depth of the β6/β7 loop upon mutation. The color scale corresponds to the plots in (A) and (B) and shows the mean number of nonpolar protein-lipid contacts per residue per ps, varying from 0 (blue) to 2 (red). Nonpolar contacts are defined as the number of POPC tail carbon atoms within 4 Å of a protein heavy atom. See also Figure S3.
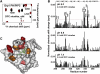
Figure 3
Association of the PI(3,4,5)P3-Bound GRP1-PH with Membrane-Mimicking DPC Micelles Monitored by NMR (A) Superimposed 1H, 15N HSQC spectra of the PI(3,4,5)P3 (0.4 mM)-bound PH domain (0.2 mM) in the DPC-free state (black) and in the presence of 5.1 mM DPC micelles (red) collected at pH 6.8. (B) The histograms show normalized (Grzesiek et al., 1996) chemical shift changes induced in the backbone amides of the PI(3,4,5)P3-bound GRP1 PH domain by DPC micelles at indicated pH. Significant changes in resonances are judged to be greater than the average plus one standard deviation (red line). (C) Residues that display significant changes in chemical shift are colored in red and pink for large and medium changes, respectively. Mutated residues are orange. The head group of PI(3,4,5)P3 is shown as a stick model and colored green.
Figure 4
Simulation Snapshots of GRP1-PH Being Pulled from the Bilayer Surface (A) Snapshot at t = 0 ns for the neutral H355 case. (B) Snapshot at t = 36 ns, again for the neutral H355 case. The interaction between the β6/β7 loop and the membrane lipids is sufficiently strong to completely remove a lipid from the bilayer (shown in yellow) at this pulling velocity, while another lipid (shown in red) is partially extracted. See also Figure S4.
Figure 5
Potential Energy of the Protein-Membrane Complex over the SMD Simulations Short-range components of the van der Waals and electrostatic contributions to the potential energy are shown for the slower SMD simulations, of duration 80 ns and a pulling rate of 0.5 Å/ns. (A) van der Waals potential energy and (B) electrostatic potential energy for the SMD simulation with H355 in a neutral state. (C) van der Waals potential energy and (D) electrostatic potential energy for the SMD simulation with H355 in a protonated state. See also Figures S1 and S2.
Figure 6
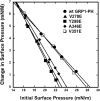
Monolayer Penetration Experiments of GRP1-PH and Respective Mutations Insertion of the wild-type GRP1 PH domain (filled circles), V278E (filled triangles), Y298E (filled squares), A346E (filled diamonds), and V351E (open squares) mutations into a POPC/POPE/PI(3,4,5)P3 (77:20:3) monolayer in a subphase of 10 mM HEPES/0.16 M KCl (pH 7.4) monitored as a function of π.
Similar articles
- Molecular mechanism of membrane binding of the GRP1 PH domain.
Lai CL, Srivastava A, Pilling C, Chase AR, Falke JJ, Voth GA. Lai CL, et al. J Mol Biol. 2013 Sep 9;425(17):3073-90. doi: 10.1016/j.jmb.2013.05.026. Epub 2013 Jun 6. J Mol Biol. 2013. PMID: 23747485 Free PMC article. - Finding a needle in a haystack: the role of electrostatics in target lipid recognition by PH domains.
Lumb CN, Sansom MS. Lumb CN, et al. PLoS Comput Biol. 2012;8(7):e1002617. doi: 10.1371/journal.pcbi.1002617. Epub 2012 Jul 26. PLoS Comput Biol. 2012. PMID: 22844242 Free PMC article. - Membrane docking geometry of GRP1 PH domain bound to a target lipid bilayer: an EPR site-directed spin-labeling and relaxation study.
Chen HC, Ziemba BP, Landgraf KE, Corbin JA, Falke JJ. Chen HC, et al. PLoS One. 2012;7(3):e33640. doi: 10.1371/journal.pone.0033640. Epub 2012 Mar 30. PLoS One. 2012. PMID: 22479423 Free PMC article. - The GRP1 PH domain, like the AKT1 PH domain, possesses a sentry glutamate residue essential for specific targeting to plasma membrane PI(3,4,5)P(3).
Pilling C, Landgraf KE, Falke JJ. Pilling C, et al. Biochemistry. 2011 Nov 15;50(45):9845-56. doi: 10.1021/bi2011306. Epub 2011 Oct 19. Biochemistry. 2011. PMID: 21932773 Free PMC article. - Molecular mechanism of membrane targeting by the GRP1 PH domain.
He J, Haney RM, Vora M, Verkhusha VV, Stahelin RV, Kutateladze TG. He J, et al. J Lipid Res. 2008 Aug;49(8):1807-15. doi: 10.1194/jlr.M800150-JLR200. Epub 2008 May 9. J Lipid Res. 2008. PMID: 18469301 Free PMC article.
Cited by
- Interactions of the auxilin-1 PTEN-like domain with model membranes result in nanoclustering of phosphatidyl inositol phosphates.
Kalli AC, Morgan G, Sansom MS. Kalli AC, et al. Biophys J. 2013 Jul 2;105(1):137-45. doi: 10.1016/j.bpj.2013.05.012. Biophys J. 2013. PMID: 23823232 Free PMC article. - Association of Peripheral Membrane Proteins with Membranes: Free Energy of Binding of GRP1 PH Domain with Phosphatidylinositol Phosphate-Containing Model Bilayers.
Naughton FB, Kalli AC, Sansom MS. Naughton FB, et al. J Phys Chem Lett. 2016 Apr 7;7(7):1219-24. doi: 10.1021/acs.jpclett.6b00153. Epub 2016 Mar 17. J Phys Chem Lett. 2016. PMID: 26977543 Free PMC article. - Defining the membrane-associated state of the PTEN tumor suppressor protein.
Lumb CN, Sansom MS. Lumb CN, et al. Biophys J. 2013 Feb 5;104(3):613-21. doi: 10.1016/j.bpj.2012.12.002. Biophys J. 2013. PMID: 23442912 Free PMC article. - Psoralen and Ultraviolet A Light Treatment Directly Affects Phosphatidylinositol 3-Kinase Signal Transduction by Altering Plasma Membrane Packing.
Van Aelst B, Devloo R, Zachée P, t'Kindt R, Sandra K, Vandekerckhove P, Compernolle V, Feys HB. Van Aelst B, et al. J Biol Chem. 2016 Nov 18;291(47):24364-24376. doi: 10.1074/jbc.M116.735126. Epub 2016 Sep 29. J Biol Chem. 2016. PMID: 27687726 Free PMC article. - Structural and Dynamic Effects of PTEN C-Terminal Tail Phosphorylation.
Smith IN, Dawson JE, Krieger J, Thacker S, Bahar I, Eng C. Smith IN, et al. J Chem Inf Model. 2022 Sep 12;62(17):4175-4190. doi: 10.1021/acs.jcim.2c00441. Epub 2022 Aug 24. J Chem Inf Model. 2022. PMID: 36001481 Free PMC article.
References
- Berendsen H.J.C., Postma J.P.M., van Gunsteren W.F., DiNola A., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690.
- Cantley L.C. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM071424/GM/NIGMS NIH HHS/United States
- GM071424/GM/NIGMS NIH HHS/United States
- BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MRC_/Medical Research Council/United Kingdom
- R01 CA095144/CA/NCI NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- CA95144/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous