The crystal structure of dynamin - PubMed (original) (raw)
The crystal structure of dynamin
Marijn G J Ford et al. Nature. 2011.
Abstract
Dynamin-related proteins (DRPs) are multi-domain GTPases that function via oligomerization and GTP-dependent conformational changes to play central roles in regulating membrane structure across phylogenetic kingdoms. How DRPs harness self-assembly and GTP-dependent conformational changes to remodel membranes is not understood. Here we present the crystal structure of an assembly-deficient mammalian endocytic DRP, dynamin 1, lacking the proline-rich domain, in its nucleotide-free state. The dynamin 1 monomer is an extended structure with the GTPase domain and bundle signalling element positioned on top of a long helical stalk with the pleckstrin homology domain flexibly attached on its opposing end. Dynamin 1 dimer and higher order dimer multimers form via interfaces located in the stalk. Analysis of these interfaces provides insight into DRP family member specificity and regulation and provides a framework for understanding the biogenesis of higher order DRP structures and the mechanism of DRP-mediated membrane scission events.
© 2011 Macmillan Publishers Limited. All rights reserved
Figures
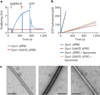
Figure 1. The G397D mutation in Dyn1 ΔPRD blocks self-assembly
. a, 90° light scattering. Dyn1 ΔPRD (blue, 1 µM) and Dyn1 G397D ΔPRD (red, 1µM) were monitored after addition of 0.5mM GMPPCP and, in the case of Dyn1 ΔPRD, 1mMGTP (blue arrows). Red arrow indicates opening of sample port. AU, arbitrary units. b, Steady-state GTP hydrolysis kinetics of Dyn1 ΔPRD and Dyn1 G397DΔPRD in the absence (light blue and red) and presence (dark blue and orange) of 0.1mgml−1 liposomes containing 10% PtdIns-4P, monitored by a NADH-dependent coupled assay as described. A representative trace is shown with 1mM GTP. c, Transmission electron microscopy of negative-stained 0.25 mgml−1 10% PtdIns-4P lipid nanotubes with Dyn1 ΔPRD (middle) and Dyn1 G397D ΔPRD (right) and no added protein (left) and 0.5mM GMPPCP. Scale bars, 200 nm.
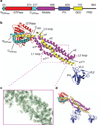
Figure 2. The crystal structure of Dyn1 G397D ΔPRD
a, Schematic of the Dyn1 domain structure. Numbers indicate domain-ending amino acid. Colour scheme used here is retained. b, Crystal structure of Dyn1 G397D ΔPRD. Linkers between the NGTPase and GTPase domain and the CGTPase and MD are shown in grey. Loops with no density are represented with dashed lines. Stalk nomenclature is based on that of the MxA stalk structure. VL, variable loop. c, An example of the refined to B-sharpened 2_mF_obs – _DF_calc map, contoured at 1s. The region shown is part of the stalk boxed by purple dotted lines in (b). d, Three symmetry-related PH domains in the lattice.
Figure 3. Dyn1 G397D ΔPRD stalk interfaces mediate self-assembly
a, Schematic diagramof fourmonomers showing interfaces in the crystal lattice. b, Surface representation showing the locations of interfaces 1 and 2. c, Detail of interface 2. Protomers are shown in lighter and darker hues. Green dotted lines are hydrogen bonds. d, Conservation of interface 2 residues involved in hydrogen bonding in dyamins, Drp1/Dnm1 and Mx proteins. Blue, conserved in dynamins, Drp1/Dnm1 and Mx proteins; yellow, conserved in dynamins and Drps; red, conserved in dynamins. Alignment shows a subset of sequences used to determine the conservation. Sequences are identical to Supplementary Fig. 1, with the addition of Rattus norvegicus Mx1 (P18588.1) and Homo sapiens Mx1 (P20591.4). Fruitfly, Drosophila melanogaster; human, Homo sapiens; rat, Rattus norvegicus; worm, Caenorhabditis elegans; yeast, Saccharomyces cerevisiae. e, Surface representation of conservation data shown in (d). f, Interface 1. Density interpreted as a PEG400 molecule is shown in black.
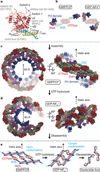
Figure 4. Oligomerization of dynamin into helical structures
a, Dynamin helices derived from the linear arrangement in our crystal structure. Two stalk dimers (green and magenta) that engage in interface 1 and 3 are related by crystallographic translation. Experimentally determined helical parameters for dynamin assembled into helices in the GMPPCP-bound state were matched by applying a small shift and tilt of one stalk dimer with respect to the other. b, Placement of oligomerized dynamin model into the electron microscopy density map contoured at 1.2s. In side view: the fit of the GTPase domain as a GTPase–GTPase dimer with the BSE in open conformation to connect to interface 1 of the stalk helix (solid density is contoured at 3.6σ). c, Observed conformational flexibility of the BSE. Model fitted into the helical reconstruction is shown as black superimposed ribbon on the crystal structure of the GTPase–CGED fusion dimer (PDB accession no. 2×2E).
Figure 5. Model for dynamin GTP cycle conformational changes
a, Mapping of dynamin shibire and sushi mutations. b, Nucleotide-dependent dynamin conformations. The GTPase core domains (red) are in the same orientation. Left, GTP-bound state with open BSE conformation of dynamin as fitted into the GMPPCP-bound electron microscopic reconstruction shown in Fig. 4. Right, transition state of dynamin obtained by superposition of the BSE residues 291–312 and 727–743 of our structure on the corresponding residues of the GDP•AlF4 −-bound GTPase–CGED fusion dimer (PDB accession no. 2×2E). Transition from open to closed BSE conformation results in movement of stalk domains. c, Model for Dyn1 ΔPRD GTP-bound helix. The BSE is opened to allow GTPase–GTPase dimer formation. d, GTP hydrolysis closes the BSE and adopts the conformation of the GDP•AlF4 −-bound transition state. This results in a substantial global constriction of the helical oligomeric assembly causing membrane deformation and scission. e, Schematic of how the proposed GTP hydrolysis triggered BSE conformational change is transmitted to oligomerized stalk domains.
Similar articles
- Crystal structure of nucleotide-free dynamin.
Faelber K, Posor Y, Gao S, Held M, Roske Y, Schulze D, Haucke V, Noé F, Daumke O. Faelber K, et al. Nature. 2011 Sep 18;477(7366):556-60. doi: 10.1038/nature10369. Nature. 2011. PMID: 21927000 - Cryo-EM of the dynamin polymer assembled on lipid membrane.
Kong L, Sochacki KA, Wang H, Fang S, Canagarajah B, Kehr AD, Rice WJ, Strub MP, Taraska JW, Hinshaw JE. Kong L, et al. Nature. 2018 Aug;560(7717):258-262. doi: 10.1038/s41586-018-0378-6. Epub 2018 Aug 1. Nature. 2018. PMID: 30069048 Free PMC article. - Crystal structure of the GTPase domain of rat dynamin 1.
Reubold TF, Eschenburg S, Becker A, Leonard M, Schmid SL, Vallee RB, Kull FJ, Manstein DJ. Reubold TF, et al. Proc Natl Acad Sci U S A. 2005 Sep 13;102(37):13093-8. doi: 10.1073/pnas.0506491102. Epub 2005 Sep 2. Proc Natl Acad Sci U S A. 2005. PMID: 16141317 Free PMC article. - Structure and function of the acidic ribosomal stalk proteins.
Wahl MC, Möller W. Wahl MC, et al. Curr Protein Pept Sci. 2002 Feb;3(1):93-106. doi: 10.2174/1389203023380756. Curr Protein Pept Sci. 2002. PMID: 12370014 Review. - The role of the PH domain and SH3 binding domains in dynamin function.
Scaife RM, Margolis RL. Scaife RM, et al. Cell Signal. 1997 Sep;9(6):395-401. doi: 10.1016/s0898-6568(97)00041-7. Cell Signal. 1997. PMID: 9376220 Review.
Cited by
- Actin bundling by dynamin 2 and cortactin is implicated in cell migration by stabilizing filopodia in human non-small cell lung carcinoma cells.
Yamada H, Takeda T, Michiue H, Abe T, Takei K. Yamada H, et al. Int J Oncol. 2016 Sep;49(3):877-86. doi: 10.3892/ijo.2016.3592. Epub 2016 Jun 30. Int J Oncol. 2016. PMID: 27572123 Free PMC article. - Dynamin-catalyzed membrane fission requires coordinated GTP hydrolysis.
Liu YW, Mattila JP, Schmid SL. Liu YW, et al. PLoS One. 2013;8(1):e55691. doi: 10.1371/journal.pone.0055691. Epub 2013 Jan 31. PLoS One. 2013. PMID: 23383266 Free PMC article. - Role of Clathrin and Dynamin in Clathrin Mediated Endocytosis/Synaptic Vesicle Recycling and Implications in Neurological Diseases.
Prichard KL, O'Brien NS, Murcia SR, Baker JR, McCluskey A. Prichard KL, et al. Front Cell Neurosci. 2022 Jan 18;15:754110. doi: 10.3389/fncel.2021.754110. eCollection 2021. Front Cell Neurosci. 2022. PMID: 35115907 Free PMC article. Review. - Dynamin photoinactivation blocks Clathrin and α-adaptin recruitment and induces bulk membrane retrieval.
Kasprowicz J, Kuenen S, Swerts J, Miskiewicz K, Verstreken P. Kasprowicz J, et al. J Cell Biol. 2014 Mar 31;204(7):1141-56. doi: 10.1083/jcb.201310090. Epub 2014 Mar 24. J Cell Biol. 2014. PMID: 24662566 Free PMC article. - Dynamin recruitment and membrane scission at the neck of a clathrin-coated pit.
Cocucci E, Gaudin R, Kirchhausen T. Cocucci E, et al. Mol Biol Cell. 2014 Nov 5;25(22):3595-609. doi: 10.1091/mbc.E14-07-1240. Epub 2014 Sep 17. Mol Biol Cell. 2014. PMID: 25232009 Free PMC article.
References
- Praefcke GJ, McMahon HT. The dynamin superfamily: universal membrane tubulation and fission molecules? Nature Rev. Mol. Cell Biol. 2004;5:133–147. - PubMed
- Hoppins S, Lackner L, Nunnari J. The machines that divide and fuse mitochondria. Annu. Rev. Biochem. 2007;76:751–780. - PubMed
- Marks B, et al. GTPase activity of dynamin and resulting conformation change are essential for endocytosis. Nature. 2001;410:231–235. - PubMed
- Stowell MHB, Marks B, Wigge P, McMahon HT. Nucleotide-dependent conformational changes in dynamin: evidence for a mechanochemical molecular spring. Nature Cell Biol. 1999;1:27–32. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01GM062942S1/GM/NIGMS NIH HHS/United States
- DRG-2004-09/HHMI/Howard Hughes Medical Institute/United States
- R01 GM097432/GM/NIGMS NIH HHS/United States
- R01 GM062942/GM/NIGMS NIH HHS/United States
- R01GM097432/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases