The deubiquitinase CYLD targets Smad7 protein to regulate transforming growth factor β (TGF-β) signaling and the development of regulatory T cells - PubMed (original) (raw)
The deubiquitinase CYLD targets Smad7 protein to regulate transforming growth factor β (TGF-β) signaling and the development of regulatory T cells
Yongge Zhao et al. J Biol Chem. 2011.
Abstract
CYLD is a lysine 63-deubiquitinating enzyme that inhibits NF-κB and JNK signaling. Here, we show that CYLD knock-out mice have markedly increased numbers of regulatory T cells (Tregs) in peripheral lymphoid organs but not in the thymus. In vitro stimulation of CYLD-deficient naive T cells with anti-CD3/28 in the presence of TGF-β led to a marked increase in the number of Foxp3-expressing T cells when compared with stimulated naive control CD4(+) cells. Under endogenous conditions, CYLD formed a complex with Smad7 that facilitated CYLD deubiquitination of Smad7 at lysine 360 and 374 residues. Moreover, this site-specific ubiquitination of Smad7 was required for activation of TAK1 and p38 kinases. Finally, knockdown of Smad7 or inhibition of p38 activity in primary T cells impaired Treg differentiation. Together, our results show that CYLD regulates TGF-β signaling function in T cells and the development of Tregs through deubiquitination of Smad7.
Figures
FIGURE 1.
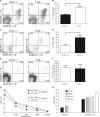
The number of Tregs is increased in the spleen of CYLD-deficient mice. A, wild type and CYLD-deficient CD4+ cells from splenocytes were analyzed for Foxp3 expression by flow cytometry. APC, allophycocyanin; PE, phycoerythrin. B, graph showing the mean and distribution of CD4+ Foxp3+ splenic T cells from wild type and CYLD-deficient mice. Twelve mice per group were analyzed. **, p < 0.01. C and D, FACS staining of mesenteric lymph nodes (MLN) performed as in A and B. E and F, FACS staining of thymocytes performed as in A and B. *, p < 0.05. Error bars represent the mean ± S.D. p values were calculated by using Student's t test. G and H, proliferative and suppressive function of CYLD-deficient CD4+ T cells. Wild type or CYLD-deficient CD4+CD25− cells and CD4+CD25+ cells, or CD4+CD25− cells co-cultured with CD4+CD25+ cells, were stimulated with antigen-presenting cells and anti-CD3. Cultures were incubated for 72 h and pulsed with [3H]TdR for the last 6 h of culture. One representative experiment of two separate experiments is shown.
FIGURE 2.
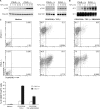
CYLD-deficient CD4 naive T cells demonstrate increased responsiveness to TGF-β in vitro. Purified wild type and CYLD-deficient CD4+CD25− T cells were cultured with TGF-β alone, in the presence of plate-bound anti-CD3/CD28 antibodies alone, or with TGF-β and anti-CD3/CD28 antibodies. At the end of the culture period, the cells were analyzed by flow cytometry. The experiment was repeated four times. Bottom panel, bar graph demonstrating the mean and distribution of CD25+ Foxp3+ T cells.
FIGURE 3.
TAK1, p38, and AP-1 activities in response to TGF-β were enhanced in CYLD-deficient CD4+ T cells. A, cellular extracts prepared from TGF-β-stimulated splenic CD4+ T cells were analyzed by immunoblotting for the expression of phospho-TAK1 (p-TAK1) and phospho-p38 (p-p38). B, cellular extracts prepared from TGF-β-stimulated splenic CD4+ T cells were subjected to EMSA for AP-1 and SP-1 binding activity. C, purified wild type and CYLD-deficient CD4+CD25− T cells were cultured with medium alone, in the presence of plate-bound anti-CD3/CD28 antibodies with TGF-β, or in the presence of anti-CD3/CD28 antibodies with TGF-β and SB 203580. At the end of the culture period, the cells were analyzed by flow cytometry. The experiment was repeated three times. Bottom panel, bar graph demonstrating the mean and distribution of CD25+ Foxp3+ T cells.
FIGURE 4.
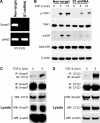
Smad7-deficient primary T cells exhibit impaired TAK1 and p38 activities. A, RT-PCR analysis of Smad7 expression in primary CD4+ T cells transduced with non-targeted or Smad7 (S7)-targeted shRNA. B, immunoblot analysis of phospho-TAK1 (p-TAK1) and phospho-p38 (p-p38) levels in TGF-β-stimulated CD4+ non-targeted cells or TGF-β-stimulated CD4+ Smad7 knockdown cells. C and D, interaction of Smad7 and CYLD in CD4+ T cells was demonstrated by immunoprecipitation (IP) of endogenous Smad7 followed by immunoblotting (IB) with antibodies against CYLD (C) or the reverse immunoprecipitation of endogenous CYLD and immunoblotting with antibodies against Smad7 (D). Western blotting of the lysates (WB) showed that equal amounts of protein were used for immunoprecipitation.
FIGURE 5.
Smad7 ubiquitination in response to TGF-β is enhanced in CYLD-deficient cells. A, Smad7 polyubiquitination in TGF-β-stimulated splenic wild type or CYLD-deficient CD4+ T cells was analyzed by immunoprecipitation (IP) of the proteins from denatured cellular lysates followed by immunoblotting (IB) with an anti-Lys-63-linked ubiquitin Ab (K63 Ub) or anti-K48-linked ubiquitin Ab (K48 Ub). The amount of immunoprecipitated Smad7 was determined by immunoblotting with anti-Smad7. WB, Western blotting; MW, molecular weight markers. B, cell lysates from HEK 293 cells transfected with pcDNA3, FLAG-Smad7, HA-ubiquitin, or HA-tagged Lys-63-only ubiquitin as indicated were immunoprecipitated as in A. The amount of polyubiquitinated FLAG-Smad7 in the immunoprecipitate was determined by immunoblotting with anti-HA, and the amount of immunoprecipitated FLAG-Smad7 was determined by immunoblotting with anti-FLAG. FLAG-Smad7 and β-actin levels in the lysate were determined by Western blotting. C, HEK 293 cells were transfected with pcDNA3, FLAG-Smad7, HA-tagged wild type ubiquitin, CYLD, and CYLD-mut lacking enzymatic activity as indicated. The cells were harvested, and the lysates were immunoprecipitated as in A. The amount of polyubiquitinated FLAG-Smad7 in the immunoprecipitate was determined by immunoblotting with anti-HA, and the amount of immunoprecipitated FLAG-Smad7 was determined by immunoblotting with anti-FLAG. The amounts of FLAG-Smad7, CYLD, and β-actin in the lysate were determined by Western blotting. D, transduced wild type and CYLD-deficient CD4+CD25− T cells were cultured with medium alone or in the presence of plate-bound anti-CD3/CD28 antibodies with TGF-β. At the end of the culture period, the cells were analyzed for CD25 and Foxp3 expression by flow cytometry. The experiment was repeated three times. Bottom left panel, bar graph demonstrating the mean and distribution of CD25+ Foxp3+ T cells. Bottom right panel, RT-PCR analysis of Smad7 expression in both WT and _Cyld_−/− primary CD4+CD25− T cells transduced with non-targeted or Smad7 (S7)-targeted shRNA.
FIGURE 6.
Lys-63-linked polyubiquitination of Smad7 at its C terminus regulates TAK1 and p38 MAPK activities in response to TGF-β. A, schematic representation of the structure of wild type Smad7 and N- and C-terminal truncation mutants. B, cell lysates were prepared from HEK 293 cells transfected with GST-tagged Smad7, GST-tagged Smad7 deletion mutants (GST-Smad7N and GST-Smad7C), and HA-tagged Lys-63-only ubiquitin (HA-UB-K63) as indicated. The lysates were subjected to immunoprecipitation (IP) as in Fig. 5_A_. The amount of polyubiquitinated GST-Smad7 in the immunoprecipitate was determined by immunoblotting (IB) with anti-HA antibody. The amounts of ubiquitin, GST-Smad7, and β-actin in the lysate were determined by Western blotting (WB). MW, molecular weight markers. C, cell lysates from HEK 293 cells transfected with FLAG-Smad7, FLAG-Smad7-K360R (Flag-S7K360R), FLAG-Smad7-K368R (Flag-S7K368R), FLAG-Smad7-K374R (Flag-S7K374R), FLAG-Smad7-K402R (Flag-S7K402R), and HA-tagged Lys-63-only ubiquitin (HA-UB-K63) were subjected to immunoprecipitation as in Fig. 5_A_. The amount of polyubiquitinated FLAG-Smad7 in the immunoprecipitate was determined by immunoblotting with anti-HA. The amounts of ubiquitin, FLAG-Smad7, and β-actin in the lysate were determined by Western blotting. D, cell lysates from HEK 293 cells transfected with FLAG-Smad7, FLAG-Smad7-K360R/K374R (Flag-S7K360/374R), FLAG-Smad7-K374R/K402R (Flag-S7K374/402R), FLAG-Smad7-K360R/K402R (Flag-S7K360/402R), and HA-tagged Lys-63-only ubiquitin (HA-Ub-K63) as indicated were harvested and immunoprecipitated as in Fig. 5_A_. The amount of polyubiquitinated FLAG-Smad7 in the immunoprecipitate was determined by immunoblotting with anti-HA. FLAG-Smad7 and β-actin in the lysate were determined by Western blotting. E, HeLa Smad7 knockdown cells were reconstituted with FLAG-Smad7 or FLAG-Smad7-K360R/K374R (K360/374R). Following TGF-β stimulation, phospho-TAK1 (p-TAK1) and phospho-p38 (p-p38) levels were determined by immunoblotting. K359/373R, K359R/K373R. F, proposed model of the role of CYLD in TGF-β signaling and Treg development. TGFR, TGF receptor; Ub, ubiquitin; P, phosphorylated.
Similar articles
- Alternative Splice Forms of CYLD Mediate Ubiquitination of SMAD7 to Prevent TGFB Signaling and Promote Colitis.
Tang Y, Reissig S, Glasmacher E, Regen T, Wanke F, Nikolaev A, Gerlach K, Popp V, Karram K, Fantini MC, Schattenberg JM, Galle PR, Neurath MF, Weigmann B, Kurschus FC, Hövelmeyer N, Waisman A. Tang Y, et al. Gastroenterology. 2019 Feb;156(3):692-707.e7. doi: 10.1053/j.gastro.2018.10.023. Epub 2018 Oct 10. Gastroenterology. 2019. PMID: 30315770 - Transforming growth factor-beta1 (TGF-beta)-induced apoptosis of prostate cancer cells involves Smad7-dependent activation of p38 by TGF-beta-activated kinase 1 and mitogen-activated protein kinase kinase 3.
Edlund S, Bu S, Schuster N, Aspenström P, Heuchel R, Heldin NE, ten Dijke P, Heldin CH, Landström M. Edlund S, et al. Mol Biol Cell. 2003 Feb;14(2):529-44. doi: 10.1091/mbc.02-03-0037. Mol Biol Cell. 2003. PMID: 12589052 Free PMC article. - CARMA1 regulation of regulatory T cell development involves modulation of interleukin-2 receptor signaling.
Lee AJ, Wu X, Cheng H, Zhou X, Cheng X, Sun SC. Lee AJ, et al. J Biol Chem. 2010 May 21;285(21):15696-703. doi: 10.1074/jbc.M109.095190. Epub 2010 Mar 16. J Biol Chem. 2010. PMID: 20233721 Free PMC article. - PKCθ/β and CYLD are antagonistic partners in the NFκB and NFAT transactivation pathways in primary mouse CD3+ T lymphocytes.
Thuille N, Wachowicz K, Hermann-Kleiter N, Kaminski S, Fresser F, Lutz-Nicoladoni C, Leitges M, Thome M, Massoumi R, Baier G. Thuille N, et al. PLoS One. 2013;8(1):e53709. doi: 10.1371/journal.pone.0053709. Epub 2013 Jan 15. PLoS One. 2013. PMID: 23335970 Free PMC article. - CYLD: a multifunctional deubiquitinase.
Glittenberg M, Ligoxygakis P. Glittenberg M, et al. Fly (Austin). 2007 Nov-Dec;1(6):330-2. doi: 10.4161/fly.5399. Epub 2007 Nov 10. Fly (Austin). 2007. PMID: 18820455 Review.
Cited by
- Deubiquitinating Enzyme: A Potential Secondary Checkpoint of Cancer Immunity.
Huang X, Zhang X, Xu J, Wang X, Zhang G, Tang T, Shen X, Liang T, Bai X. Huang X, et al. Front Oncol. 2020 Aug 7;10:1289. doi: 10.3389/fonc.2020.01289. eCollection 2020. Front Oncol. 2020. PMID: 32850399 Free PMC article. Review. - Reversal of prolonged obesity-associated cerebrovascular dysfunction by inhibiting microglial Tak1.
Shen Q, Chen Z, Zhao F, Pan S, Zhang T, Cheng X, Zhang L, Zhang S, Qi J, Li J, Cai D, Zhang G. Shen Q, et al. Nat Neurosci. 2020 Jul;23(7):832-841. doi: 10.1038/s41593-020-0642-6. Epub 2020 May 25. Nat Neurosci. 2020. PMID: 32451485 - De-ubiquitinating enzyme, USP11, promotes transforming growth factor β-1 signaling through stabilization of transforming growth factor β receptor II.
Jacko AM, Nan L, Li S, Tan J, Zhao J, Kass DJ, Zhao Y. Jacko AM, et al. Cell Death Dis. 2016 Nov 17;7(11):e2474. doi: 10.1038/cddis.2016.371. Cell Death Dis. 2016. PMID: 27853171 Free PMC article. - Molecular Mechanisms of DUBs Regulation in Signaling and Disease.
Li Y, Reverter D. Li Y, et al. Int J Mol Sci. 2021 Jan 20;22(3):986. doi: 10.3390/ijms22030986. Int J Mol Sci. 2021. PMID: 33498168 Free PMC article. Review. - CYLD, a mechanosensitive deubiquitinase, regulates TGFβ signaling in load-induced bone formation.
Nguyen J, Massoumi R, Alliston T. Nguyen J, et al. Bone. 2020 Feb;131:115148. doi: 10.1016/j.bone.2019.115148. Epub 2019 Nov 9. Bone. 2020. PMID: 31715338 Free PMC article.
References
- Sakaguchi S., Yamaguchi T., Nomura T., Ono M. (2008) Cell 133, 775–787 - PubMed
- Hori S., Nomura T., Sakaguchi S. (2003) Science 299, 1057–1061 - PubMed
- Fontenot J. D., Gavin M. A., Rudensky A. Y. (2003) Nat. Immunol. 4, 330–336 - PubMed
- Khattri R., Cox T., Yasayko S. A., Ramsdell F. (2003) Nat. Immunol. 4, 337–342 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous