Discovery of the first insect nidovirus, a missing evolutionary link in the emergence of the largest RNA virus genomes - PubMed (original) (raw)
. 2011 Sep;7(9):e1002215.
doi: 10.1371/journal.ppat.1002215. Epub 2011 Sep 8.
Maria del Carmen Parquet, Chris Lauber, Manmohan Parida, Takeshi Nabeshima, Fuxun Yu, Nguyen Thanh Thuy, Shingo Inoue, Takashi Ito, Kenta Okamoto, Akitoyo Ichinose, Eric J Snijder, Kouichi Morita, Alexander E Gorbalenya
Affiliations
- PMID: 21931546
- PMCID: PMC3169540
- DOI: 10.1371/journal.ppat.1002215
Discovery of the first insect nidovirus, a missing evolutionary link in the emergence of the largest RNA virus genomes
Phan Thi Nga et al. PLoS Pathog. 2011 Sep.
Abstract
Nidoviruses with large genomes (26.3-31.7 kb; 'large nidoviruses'), including Coronaviridae and Roniviridae, are the most complex positive-sense single-stranded RNA (ssRNA+) viruses. Based on genome size, they are far separated from all other ssRNA+ viruses (below 19.6 kb), including the distantly related Arteriviridae (12.7-15.7 kb; 'small nidoviruses'). Exceptionally for ssRNA+ viruses, large nidoviruses encode a 3'-5'exoribonuclease (ExoN) that was implicated in controlling RNA replication fidelity. Its acquisition may have given rise to the ancestor of large nidoviruses, a hypothesis for which we here provide evolutionary support using comparative genomics involving the newly discovered first insect-borne nidovirus. This Nam Dinh virus (NDiV), named after a Vietnamese province, was isolated from mosquitoes and is yet to be linked to any pathology. The genome of this enveloped 60-80 nm virus is 20,192 nt and has a nidovirus-like polycistronic organization including two large, partially overlapping open reading frames (ORF) 1a and 1b followed by several smaller 3'-proximal ORFs. Peptide sequencing assigned three virion proteins to ORFs 2a, 2b, and 3, which are expressed from two 3'-coterminal subgenomic RNAs. The NDiV ORF1a/ORF1b frameshifting signal and various replicative proteins were tentatively mapped to canonical positions in the nidovirus genome. They include six nidovirus-wide conserved replicase domains, as well as the ExoN and 2'-O-methyltransferase that are specific to large nidoviruses. NDiV ORF1b also encodes a putative N7-methyltransferase, identified in a subset of large nidoviruses, but not the uridylate-specific endonuclease that - in deviation from the current paradigm - is present exclusively in the currently known vertebrate nidoviruses. Rooted phylogenetic inference by Bayesian and Maximum Likelihood methods indicates that NDiV clusters with roniviruses and that its branch diverged from large nidoviruses early after they split from small nidoviruses. Together these characteristics identify NDiV as the prototype of a new nidovirus family and a missing link in the transition from small to large nidoviruses.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Figure 1. Distribution of positive-sense single-stranded RNA virus genome sizes.
The Coronaviridae is split into the corona- and toro-/bafinivirus groups. Prefix Nido- and Picorna- are for Nidovirales and Picornavirales, respectively. Group specific box-whisker plots are aligned along the X-axis by their medians (bold line) normalized to zero. The box spans from the first to third quartile; the whiskers (dashed lines) are <1.5 times the inter-quartile range; outliers are circles. Families/groups are ranked by median genome size along Y-axis. Genome size ranges are colored by semi-circles: nidoviruses (dark orange), other classified (dark grey) and unclassified viruses (light grey). Three non-overlapping zones regarding the presence of the exoribonuclease (ExoN) are highlighted in the genome size distribution (from top to bottom): ExoN-encoding large nidoviruses and NDiV (light green); in-between not-sampled size zone (white); ExoN-lacking ssRNA+ viruses (light blue).
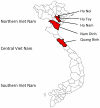
Figure 2. Map of the Vietnam provinces, where mosquito surveillance was conducted between 2001 and 2003.
Figure 3. NDiV characteristics.
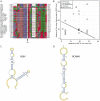
(A) Electron micrograph of negatively stained NDiV virions. (B) SDS-PAGE analysis of NDiV virion proteins. (C) Detection of NDiV genomic (mRNA1) and subgenomic mRNAs (mRNAs 2 and 3) by Northern blot hybridization analysis of total intracellular RNA from virus-infected cells, using a radiolabeled probe complementary to the 3′end of the NDiV genome. (D) NDiV genome organization and expression. Open reading frames (ORFs) are represented by open rectangles and ORF1a- and ORF1b-encoded protein domains identified by bioinformatics analyses (see Table 2) are highlighted in grey. Peptide sequences of virion proteins were determined as described in the Materials and Methods section and mapped to the products of ORFs 2a, 2b, and 3 (bottom-right). N-terminal protein sequences are indicated by (*), other peptide sequences indicate inner sequences. The actual molecular size of the ORF2a product (approximately 77 KDa in SDS-PAGE in B) is considerably shorter than the calculated size (102 KDa), suggesting that p2a may be post-translationally proteolytically processed or that its translation starts at another AUG codn in the ORF. Two pairs of conserved potential TRSs – for sg mRNAs 2 and 3, respectively - were identified in the NDiV genome and aligned (bottom-left), with each pair consisting of a putative leader TRS in the 5′-UTR and a body TRS in the 3′-proximal region of the genome. Between these TRS pairs, eight and three positions include complete match (*) and nucleotide overlap (:), respectively.
Figure 4. ORF1a/ORF1b ribosomal frameshifting in the NDiV genome.
(A) The nucleotide alignment around the open reading frame (ORF) 1a/1b ribosomal −1 frameshift (RFS) in 28 nidovirus species. The alignment column marked with an arrow indicates the RFS position and precedes a nucleotide which is read twice, as the third nucleotide of an ORF1a codon and the first coding nucleotide of ORF1b, respectively. In NDiV this residue is predicted to be located at genome position 7851. Numbers flanking the alignment indicate genomic positions. (B) RFS position in the ORF1a/ORF1b overlap of nidoviruses. The RFS position in selected nidoviruses is plotted as distances between RFS and termination codons flanking ORF1b and ORF1a from upstream and downstream, respectively. In NDiV, the predicted RFS (filled triangle) and all other candidate positions are located on black line in the 40 nt ORF1a/ORF1b overlap. (C and D) RNA secondary structure of sequence fragments around of the ORF1a/ORF1b overlap in NDiV and Red clover necrotic mosaic virus (RCNMV) as predicted by pknotsRG . The putative slippery sequences are in red (see also A). The predicted stem loop in RCNMV closely matches the one presented in .
Figure 5. Alignments of ExoN, OMT and NMT domains of NDiV and other nidoviruses.
Alignments were compiled utilizing the Muscle program followed by manual inspection. Pictures by JalView and residues are colored according to degree of conservation. Numbers above a column indicate its absolute position in the alignment (start = 1); numbers to the left and to the right of the alignment represent positions in the genome. Selected conserved sequence motifs are highlighted with black bars and roman numbers. (A) In the exonuclease (ExoN) alignment, three motifs are part of the catalytic centre; the domain includes two putative zinc fingers, specific either for roniviruses or for all nidoviruses and highlighted by, respectively, red and green asterisks. (B) In the 2′-_O_-methyltransferase (OMT) alignment, motifs X, IV, VI and VIII include residues of the catalytic tetrad (KDKE, marked with green asterisks) and motif I is involved in binding of the methyl donor . (C) Protein secondary structure predictions by Psipred for the profiles of N7-methyltransferase (NMT) from 3 NDiV/roniviruses (pred1) and 17 coronaviruses (pred2) and corresponding confidence values (conf1, conf2) were added above the alignment. Only 3 coronaviruses, representing alpha- (HCoV NL63), beta- (SARS-CoV) and gammacoronaviruses (IBV), are shown that results in several empty alignment columns. The black bar on top is a region including the methyl-donor binding site (motif I, delineated by [49]) that gave a hit with a functionally similar site of a cellular guanine N7-methyltransferase (fungus Encephalitozoon cuniculi) upon HH search of the SCOP database (data not shown). Green asterisks, conserved Cys/His residues that may form a zinc finger.
Figure 6. Phylogeny of nidoviruses.
To infer phylogenetic relationships between NDiV and other nidoviruses partially constrained trees were calculated using either a concatenated alignment of the three nidovirus-wide conserved domains (A and B) or one nidovirus-wide conserved domain (C and D). For all trees, internal nodes without support value that were fixed prior to the analysis are marked with *, otherwise, numbers indicate posterior probability support values (at the scale from 0 to 1) obtained in either (sub)family- or order-wide analyses (grey and black, respectively). The tree scale bars represent number of substitutions per amino acid position on average. (A) and (B), trees with the constrained topology in which coronaviruses and toro-/bafiniviruses were either fixed as sister clades or not, respectively. Shown are trees for the original alignment which were similar to those obtained for the alignment derivative in which least conserved columns were removed (see Materials and Methods). The trees were rooted on the arterivirus branch. (C) and (D), trees based on conserved HEL1 and RdRp domains, respectively, and including a domain-specific outgroup as described in Materials and Methods. The sister position of coronaviruses and toro-/bafiniviruses was not fixed. For virus listing see trees in A and B. Support values for the outgroup branching in a Maximum-Likelihood analysis with 1000 non-parametric bootstraps, which resulted in an identical topology, is shown below posterior probability support values in both trees. Support values for internal branching within the Coronavirinae subfamily and the Arteriviridae family are omitted for clarity. The outgroup placement on the arterivirus branch in these analyses was used to root trees in A and B.
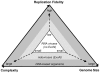
Figure 7. The Eigen trap and a model of nidoviral escape by ExoN acquisition.
The scheme depicts the unidirectional relationship between replication fidelity, genome size, and complexity. The vector of variation for the dimensions defined by the three elements of the relationship is shown in a simplified form. The position of RNA viruses in the white triangular space (Eigen trap) and the proposed effect of ExoN acquisition in nidoviruses on this position are indicated.
Similar articles
- A planarian nidovirus expands the limits of RNA genome size.
Saberi A, Gulyaeva AA, Brubacher JL, Newmark PA, Gorbalenya AE. Saberi A, et al. PLoS Pathog. 2018 Nov 1;14(11):e1007314. doi: 10.1371/journal.ppat.1007314. eCollection 2018 Nov. PLoS Pathog. 2018. PMID: 30383829 Free PMC article. - Characterization of White bream virus reveals a novel genetic cluster of nidoviruses.
Schütze H, Ulferts R, Schelle B, Bayer S, Granzow H, Hoffmann B, Mettenleiter TC, Ziebuhr J. Schütze H, et al. J Virol. 2006 Dec;80(23):11598-609. doi: 10.1128/JVI.01758-06. Epub 2006 Sep 20. J Virol. 2006. PMID: 16987966 Free PMC article. - Giant RNA genomes: Roles of host, translation elongation, genome architecture, and proteome in nidoviruses.
Neuman BW, Smart A, Gilmer O, Smyth RP, Vaas J, Böker N, Samborskiy DV, Bartenschlager R, Seitz S, Gorbalenya AE, Caliskan N, Lauber C. Neuman BW, et al. Proc Natl Acad Sci U S A. 2025 Feb 18;122(7):e2413675122. doi: 10.1073/pnas.2413675122. Epub 2025 Feb 10. Proc Natl Acad Sci U S A. 2025. PMID: 39928875 - Nidovirales: evolving the largest RNA virus genome.
Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ. Gorbalenya AE, et al. Virus Res. 2006 Apr;117(1):17-37. doi: 10.1016/j.virusres.2006.01.017. Epub 2006 Feb 28. Virus Res. 2006. PMID: 16503362 Free PMC article. Review. - Nidovirus ribonucleases: Structures and functions in viral replication.
Ulferts R, Ziebuhr J. Ulferts R, et al. RNA Biol. 2011 Mar-Apr;8(2):295-304. doi: 10.4161/rna.8.2.15196. Epub 2011 Mar 1. RNA Biol. 2011. PMID: 21422822 Review.
Cited by
- Cyclophilins and cyclophilin inhibitors in nidovirus replication.
de Wilde AH, Pham U, Posthuma CC, Snijder EJ. de Wilde AH, et al. Virology. 2018 Sep;522:46-55. doi: 10.1016/j.virol.2018.06.011. Epub 2018 Jul 12. Virology. 2018. PMID: 30014857 Free PMC article. Review. - A meta-transcriptomic study of mosquito virome and blood feeding patterns at the human-animal-environment interface in Guangdong Province, China.
Wu Q, Guo C, Li XK, Yi BY, Li QL, Guo ZM, Lu JH. Wu Q, et al. One Health. 2023 Feb 1;16:100493. doi: 10.1016/j.onehlt.2023.100493. eCollection 2023 Jun. One Health. 2023. PMID: 36817976 Free PMC article. - Replication of the coronavirus genome: A paradox among positive-strand RNA viruses.
Grellet E, L'Hôte I, Goulet A, Imbert I. Grellet E, et al. J Biol Chem. 2022 May;298(5):101923. doi: 10.1016/j.jbc.2022.101923. Epub 2022 Apr 10. J Biol Chem. 2022. PMID: 35413290 Free PMC article. Review. - Increasing the number of available ranks in virus taxonomy from five to ten and adopting the Baltimore classes as taxa at the basal rank.
Gorbalenya AE. Gorbalenya AE. Arch Virol. 2018 Oct;163(10):2933-2936. doi: 10.1007/s00705-018-3915-6. Epub 2018 Jun 26. Arch Virol. 2018. PMID: 29942981 Free PMC article. - Comparative Metagenomic Profiling of Viromes Associated with Four Common Mosquito Species in China.
Xia H, Wang Y, Shi C, Atoni E, Zhao L, Yuan Z. Xia H, et al. Virol Sin. 2018 Feb;33(1):59-66. doi: 10.1007/s12250-018-0015-4. Epub 2018 Mar 2. Virol Sin. 2018. PMID: 29500689 Free PMC article.
References
- Jenkins GM, Rambaut A, Pybus OG, Holmes EC. Rates of molecular evolution in RNA viruses: A quantitative phylogenetic analysis. J Mol Evol. 2002;54:156–165. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials