Dbp5, Gle1-IP6 and Nup159: a working model for mRNP export - PubMed (original) (raw)
Review
Dbp5, Gle1-IP6 and Nup159: a working model for mRNP export
Andrew W Folkmann et al. Nucleus. 2011 Nov-Dec.
Abstract
Gene expression is a stepwise process involving distinct cellular processes including transcription, mRNA (mRNA) processing, mRNA export, and translation. As mRNAs are being synthesized, proteins associate with the RNA to form messenger ribonucleoprotein particles (mRNPs). Previous studies have demonstrated that the RNA-binding protein composition of these mRNPs is dynamic, changing as the mRNP moves through the different steps of gene expression, and playing a critical role in these events. An important step during this maturation process occurs at the cytoplasmic face of the nuclear pore complex (NPC) where the export protein Gle1 bound to inositol hexakisphosphate (IP 6) spatially activates the ATP-hydrolysis and mRNP-remodeling activity of the DEAD-box protein Dbp5. Recent work from our laboratory and others has provided important insights into the function and regulation of Dbp5. These include a more detailed explanation of the mechanism of Dbp5 RNP remodeling, the role of Gle1-IP6 in stimulating Dbp5 ATPase activity, and the identification of a novel paradigm for regulation of Dbp5 by Nup159. Based on in vitro biochemical assays, X-ray crystallography, and corresponding in vivo phenotypes, we propose here an updated model of the Dbp5 cycle during mRNP export through the NPC. This takes into account all available data and provides a platform for future studies.
Figures
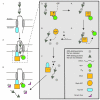
Figure 1. Structural organization of a DEAD box protein. (A) Topology model of the RecA-like helicase domains of a generic DEAD-box protein. The positions of the conserved sequence motifs within the labeled RecA helicase domains are indicated with Roman numerals. (B) Structure of human Dbp5 bound with ATP analog (black) [PDB 3GOH]. Highlighted conserved sequence motifs include those involved in nucleotide binding and hydrolysis (Green); RNA binding and ATP hydrolysis (Pink); RNA binding (Blue).
Figure 3. Working model for Dbp5/Gle1-IP6/Nup159 mRNA export cycle. During export, the mRNP exits the NPC where it encounters both Gle1-IP6 and Dbp5 at the cytoplasmic fibrils (Steps 1–2). Gle1-IP6 binds Dbp5 and enhances ATP loading (Steps 3–4). The ATP/Dbp5/Gle1-IP6 complex then binds to the mRNP, which stimulates both the release of Gle1-IP6 and the ATP hydrolysis event (Step 5). The change from ATP to ADP triggers a conformational change that drives both the remodeling of the mRNP and release of the mRNA from Dbp5 (Step 6). Dbp5-ADP is then recycled by interaction with Nup159 to release ADP (Step 7), and positioning for binding to Gle1 to begin the cycle again. The released RNA-binding proteins bind to cytoplasmic karyopherins for import back into the nucleus (Step 8).
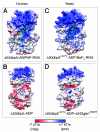
Figure 2. Solvent electrostatics of S. cerevisiae and human Dbp5. Solvent-excluded surface views of (A) human Δ53dbp5-ANPNP-RNA protein [PDB 3G0H], (B) human Δ53Dbp5-ADP protein [PDB 3EWS], (C) yeast (S. cerevisiae) Δ90dbp5L327V-ADP·BeF3 protein [PDB 3PEW], and (D) yeast (S. cerevisiae) Δ90dbp5L327V-ADP-Δ243 gle1H337R_-_IP6 proteins [PDB 3RRN] are shown colored by solvent accessible electrostatics contoured at −7 (red) to +7 _k_T/e (blue). The electrostatic potential was mapped to a molecular surface calculated using UCSF Chimera with default settings, as indicated by the PDB coordinates in brackets above. RNA is shown in green.
Comment on
- Hodge CA, Tran EJ, Noble KN, Alcazar-Roman AR, Ben-Yishay R, Scarcelli JJ, Folkmann AW, Shav-Tal Y, Wente SR, Cole CN. The Dbp5 cycle at the nuclear pore complex during mRNA export I: dbp5 mutants with defects in RNA binding and ATP hydrolysis define key steps for Nup159 and Gle1. Genes Dev. 2011;25 doi: 10.1101/gad.2041611.
- Noble KN, Tran EJ, Alcázar-Román AR, Hodge CA, Cole CN, Wente SR. The Dbp5 cycle at the nuclear pore complex during mRNA export II: nucleotide cycling and mRNP remodeling by Dbp5 are controlled by Nup159 and Gle1. Genes Dev. 2011;25 doi: 10.1101/gad.2040611.
Similar articles
- The Dbp5 cycle at the nuclear pore complex during mRNA export II: nucleotide cycling and mRNP remodeling by Dbp5 are controlled by Nup159 and Gle1.
Noble KN, Tran EJ, Alcázar-Román AR, Hodge CA, Cole CN, Wente SR. Noble KN, et al. Genes Dev. 2011 May 15;25(10):1065-77. doi: 10.1101/gad.2040611. Genes Dev. 2011. PMID: 21576266 Free PMC article. - The Dbp5 cycle at the nuclear pore complex during mRNA export I: dbp5 mutants with defects in RNA binding and ATP hydrolysis define key steps for Nup159 and Gle1.
Hodge CA, Tran EJ, Noble KN, Alcazar-Roman AR, Ben-Yishay R, Scarcelli JJ, Folkmann AW, Shav-Tal Y, Wente SR, Cole CN. Hodge CA, et al. Genes Dev. 2011 May 15;25(10):1052-64. doi: 10.1101/gad.2041611. Genes Dev. 2011. PMID: 21576265 Free PMC article. - A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export.
Montpetit B, Thomsen ND, Helmke KJ, Seeliger MA, Berger JM, Weis K. Montpetit B, et al. Nature. 2011 Apr 14;472(7342):238-42. doi: 10.1038/nature09862. Epub 2011 Mar 27. Nature. 2011. PMID: 21441902 Free PMC article. - Dbp5 - from nuclear export to translation.
Tieg B, Krebber H. Tieg B, et al. Biochim Biophys Acta. 2013 Aug;1829(8):791-8. doi: 10.1016/j.bbagrm.2012.10.010. Epub 2012 Nov 2. Biochim Biophys Acta. 2013. PMID: 23128325 Review. - Ratcheting mRNA out of the nucleus.
Stewart M. Stewart M. Mol Cell. 2007 Feb 9;25(3):327-30. doi: 10.1016/j.molcel.2007.01.016. Mol Cell. 2007. PMID: 17289581 Review.
Cited by
- Aiding and abetting cancer: mRNA export and the nuclear pore.
Culjkovic-Kraljacic B, Borden KL. Culjkovic-Kraljacic B, et al. Trends Cell Biol. 2013 Jul;23(7):328-35. doi: 10.1016/j.tcb.2013.03.004. Epub 2013 Apr 10. Trends Cell Biol. 2013. PMID: 23582887 Free PMC article. Review. - DEAD-ly Affairs: The Roles of DEAD-Box Proteins on HIV-1 Viral RNA Metabolism.
Rao S, Mahmoudi T. Rao S, et al. Front Cell Dev Biol. 2022 Jun 13;10:917599. doi: 10.3389/fcell.2022.917599. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35769258 Free PMC article. Review. - Temporal and spatial regulation of mRNA export: Single particle RNA-imaging provides new tools and insights.
Heinrich S, Derrer CP, Lari A, Weis K, Montpetit B. Heinrich S, et al. Bioessays. 2017 Feb;39(2):10.1002/bies.201600124. doi: 10.1002/bies.201600124. Epub 2017 Jan 4. Bioessays. 2017. PMID: 28052353 Free PMC article. Review. - The Proteasome Activator PA200/PSME4: An Emerging New Player in Health and Disease.
Yazgili AS, Ebstein F, Meiners S. Yazgili AS, et al. Biomolecules. 2022 Aug 20;12(8):1150. doi: 10.3390/biom12081150. Biomolecules. 2022. PMID: 36009043 Free PMC article. Review. - Insights into mRNA export-linked molecular mechanisms of human disease through a Gle1 structure-function analysis.
Folkmann AW, Dawson TR, Wente SR. Folkmann AW, et al. Adv Biol Regul. 2014 Jan;54:74-91. doi: 10.1016/j.jbior.2013.10.002. Epub 2013 Nov 13. Adv Biol Regul. 2014. PMID: 24275432 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- T32 CA119925/CA/NCI NIH HHS/United States
- GM33998/GM/NIGMS NIH HHS/United States
- F31 HD061181/HD/NICHD NIH HHS/United States
- P30 CA023108/CA/NCI NIH HHS/United States
- 1F31 HD061181/HD/NICHD NIH HHS/United States
- 1F31 NS070431/NS/NINDS NIH HHS/United States
- R37 GM051219/GM/NIGMS NIH HHS/United States
- R01 GM033998/GM/NIGMS NIH HHS/United States
- R37 GM51219/GM/NIGMS NIH HHS/United States
- F31 NS070431/NS/NINDS NIH HHS/United States
- P41 RR001081/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials