ReCount: a multi-experiment resource of analysis-ready RNA-seq gene count datasets - PubMed (original) (raw)
ReCount: a multi-experiment resource of analysis-ready RNA-seq gene count datasets
Alyssa C Frazee et al. BMC Bioinformatics. 2011.
Abstract
Background: RNA sequencing is a flexible and powerful new approach for measuring gene, exon, or isoform expression. To maximize the utility of RNA sequencing data, new statistical methods are needed for clustering, differential expression, and other analyses. A major barrier to the development of new statistical methods is the lack of RNA sequencing datasets that can be easily obtained and analyzed in common statistical software packages such as R. To speed up the development process, we have created a resource of analysis-ready RNA-sequencing datasets. 2 DESCRIPTION: ReCount is an online resource of RNA-seq gene count tables and auxilliary data. Tables were built from raw RNA sequencing data from 18 different published studies comprising 475 samples and over 8 billion reads. Using the Myrna package, reads were aligned, overlapped with gene models and tabulated into gene-by-sample count tables that are ready for statistical analysis. Count tables and phenotype data were combined into Bioconductor ExpressionSet objects for ease of analysis. ReCount also contains the Myrna manifest files and R source code used to process the samples, allowing statistical and computational scientists to consider alternative parameter values. 3 CONCLUSIONS: By combining datasets from many studies and providing data that has already been processed from. fastq format into ready-to-use. RData and. txt files, ReCount facilitates analysis and methods development for RNA-seq count data. We anticipate that ReCount will also be useful for investigators who wish to consider cross-study comparisons and alternative normalization strategies for RNA-seq.
Figures
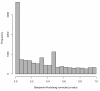
Figure 1
Histogram of adjusted p-values from differential expression analysis on the 29 samples included in both Cheung and Montgomery. The p-values in the histogram are from paired t-tests on the 25% of genes with nonzero counts in at least one of the two studies. The peak near zero is somewhat indicative of technical variability between the two studies.
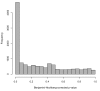
Figure 2
Histogram of adjusted p-values from analysis of differential expression between YRI and CEU populations. The p-values in the histogram are from two-sample t-tests on the 25% of genes with nonzero counts in at least one of the two studies. The peak near zero indicates differential gene expression that may result from either technical or biological variability.
Similar articles
- Cloud-scale RNA-sequencing differential expression analysis with Myrna.
Langmead B, Hansen KD, Leek JT. Langmead B, et al. Genome Biol. 2010;11(8):R83. doi: 10.1186/gb-2010-11-8-r83. Epub 2010 Aug 11. Genome Biol. 2010. PMID: 20701754 Free PMC article. - JingleBells: A Repository of Immune-Related Single-Cell RNA-Sequencing Datasets.
Ner-Gaon H, Melchior A, Golan N, Ben-Haim Y, Shay T. Ner-Gaon H, et al. J Immunol. 2017 May 1;198(9):3375-3379. doi: 10.4049/jimmunol.1700272. J Immunol. 2017. PMID: 28416714 - TCC: an R package for comparing tag count data with robust normalization strategies.
Sun J, Nishiyama T, Shimizu K, Kadota K. Sun J, et al. BMC Bioinformatics. 2013 Jul 9;14:219. doi: 10.1186/1471-2105-14-219. BMC Bioinformatics. 2013. PMID: 23837715 Free PMC article. - Differential Expression Analysis of RNA-seq Reads: Overview, Taxonomy, and Tools.
Chowdhury HA, Bhattacharyya DK, Kalita JK. Chowdhury HA, et al. IEEE/ACM Trans Comput Biol Bioinform. 2020 Mar-Apr;17(2):566-586. doi: 10.1109/TCBB.2018.2873010. Epub 2018 Oct 1. IEEE/ACM Trans Comput Biol Bioinform. 2020. PMID: 30281477 Review. - Normalization for Single-Cell RNA-Seq Data Analysis.
Bacher R. Bacher R. Methods Mol Biol. 2019;1935:11-23. doi: 10.1007/978-1-4939-9057-3_2. Methods Mol Biol. 2019. PMID: 30758817 Review.
Cited by
- Expression profile of ectopic olfactory receptors determined by deep sequencing.
Flegel C, Manteniotis S, Osthold S, Hatt H, Gisselmann G. Flegel C, et al. PLoS One. 2013;8(2):e55368. doi: 10.1371/journal.pone.0055368. Epub 2013 Feb 6. PLoS One. 2013. PMID: 23405139 Free PMC article. - An Iterative Leave-One-Out Approach to Outlier Detection in RNA-Seq Data.
George NI, Bowyer JF, Crabtree NM, Chang CW. George NI, et al. PLoS One. 2015 Jun 3;10(6):e0125224. doi: 10.1371/journal.pone.0125224. eCollection 2015. PLoS One. 2015. PMID: 26039068 Free PMC article. - Elucidating tissue specific genes using the Benford distribution.
Karthik D, Stelzer G, Gershanov S, Baranes D, Salmon-Divon M. Karthik D, et al. BMC Genomics. 2016 Aug 9;17:595. doi: 10.1186/s12864-016-2921-x. BMC Genomics. 2016. PMID: 27506195 Free PMC article. - Sparse sliced inverse regression for high dimensional data analysis.
Hilafu H, Safo SE. Hilafu H, et al. BMC Bioinformatics. 2022 May 7;23(1):168. doi: 10.1186/s12859-022-04700-3. BMC Bioinformatics. 2022. PMID: 35525975 Free PMC article. - Differential abundance analysis for microbial marker-gene surveys.
Paulson JN, Stine OC, Bravo HC, Pop M. Paulson JN, et al. Nat Methods. 2013 Dec;10(12):1200-2. doi: 10.1038/nmeth.2658. Epub 2013 Sep 29. Nat Methods. 2013. PMID: 24076764 Free PMC article.
References
- Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Muertter RN, Holko M, Ayanbule O, Yefanov A, Sobolera A. NCBI GEO: archive for functional genomics data sets - 10 years on. Nucleic Acids Res. 2011;39(suppl 1):D1005–D1010. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HG005220/HG/NHGRI NIH HHS/United States
- T32GM074906/GM/NIGMS NIH HHS/United States
- P41-HG004059/HG/NHGRI NIH HHS/United States
- R01-HG005220/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources