Cdh1 regulates osteoblast function through an APC/C-independent modulation of Smurf1 - PubMed (original) (raw)
. 2011 Dec 9;44(5):721-33.
doi: 10.1016/j.molcel.2011.09.024.
Weiguo Zou, Daming Gao, Hiroyuki Inuzuka, Hidefumi Fukushima, Anders H Berg, Rebecca Drapp, Shavali Shaik, Dorothy Hu, Chantel Lester, Manuel Eguren, Marcos Malumbres, Laurie H Glimcher, Wenyi Wei
Affiliations
- PMID: 22152476
- PMCID: PMC3240853
- DOI: 10.1016/j.molcel.2011.09.024
Cdh1 regulates osteoblast function through an APC/C-independent modulation of Smurf1
Lixin Wan et al. Mol Cell. 2011.
Abstract
The APC/Cdh1 E3 ubiquitin ligase plays an essential role in both mitotic exit and G1/S transition by targeting key cell-cycle regulators for destruction. There is mounting evidence indicating that Cdh1 has other functions in addition to cell-cycle regulation. However, it remains unclear whether these additional functions depend on its E3 ligase activity. Here, we report that Cdh1, but not Cdc20, promotes the E3 ligase activity of Smurf1. This is mediated by disruption of an autoinhibitory Smurf1 homodimer and is independent of APC/Cdh1 E3 ligase activity. As a result, depletion of Cdh1 leads to reduced Smurf1 activity and subsequent activation of multiple downstream targets, including the MEKK2 signaling pathway, inducing osteoblast differentiation. Our studies uncover a cell-cycle-independent function of Cdh1, establishing Cdh1 as an upstream component that governs Smurf1 activity. They further suggest that modulation of Cdh1 is a potential therapeutic option for treatment of osteoporosis.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
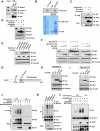
Figure 1. Cdh1 interacts with Smurf1 to promote Smurf1 auto-ubiquitination
A. Immunoblot (IB) analysis of 293T whole cell lysates (WCL) and anti-Cdh1 immunoprecipitates (IP). Mouse IgG was used as a negative control for the immunoprecipitation procedure. B. Autoradiography of 35S-labelled Smurf1 bound to the indicated GST fusion proteins. C. Immunoblot (IB) analysis of whole cell lysates (WCL) and immunoprecipitates (IP) derived from 293T cells transfected with HA-Cdh1 or HA-Cdc20 and Flag-Smurf1 constructs. Thirty hours post-transfection, cells were pretreated with 10 μM MG132 for 10 hours to block the proteasome pathway before harvesting. D. Immunoblot (IB) analysis of whole cell lysates (WCL) derived from 293T cells transfected with HA-Cdh1 or HA-Cdc20 together with Myc-tagged Smurf1 constructs. Where indicated, 10 μM MG132 was added for 10 hours to block the proteasome pathway before harvesting. E. Immunoblot (IB) analysis of whole cell lysates (WCL) derived from 293T cells transfected with HA-Cdh1 and the indicated Myc-tagged Smurf1 or Smurf2 constructs. CA-Smurf1 is an E3 ligase activity-deficient mutant form of Smurf1 in which the cysteine residue in the active site is replaced with alanine. F. Immunoblot (IB) analysis of whole cell lysates (WCL) derived from 293T cells transfected with Myc-tagged Smurf1 or CA-Smurf1 in the presence of increasing amounts of HA-Cdh1. Where indicated, 10 μM MG132 was added for 10 hours to block the proteasome pathway before harvesting. G. Schematic representation of the Cdh1 protein functional domains. H. Immunoblot (IB) analysis of whole cell lysates (WCL) derived from 293T cells transfected with Myc-Smurf1 and the indicated HA-Cdh1 constructs. I. Immunoblot (IB) analysis of whole cell lysates (WCL) derived from HeLa cells transfected with Flag-Cyclin B1 and the indicated HA-Cdh1 constructs. J. Immunoblot analysis (IB) of whole cell lysates (WCL) and anti-Flag immunoprecipitates (IP) derived from 293T cells transfected with the indicated plasmids. Twenty hours post-transfection, cells were treated with the proteasome inhibitor MG132 overnight before harvesting. K. Immunoblot analysis (IB) of whole cell lysates (WCL) and anti-Smurf1 immunoprecipitates (IP) derived from HCT116 cells infected with the indicated lentiviral shRNA constructs. Twenty hours post-infection, cells were selected with 1 μg/ml puromycin for 72 hours to eliminate non-infected cells. Cells were treated with the proteasome inhibitor MG132 overnight before harvesting. L. Purified recombinant GST-Cdh1 augments Smurf1 auto-ubiquitination in vitro. Bacterially expressed and purified GST-Smurf1 was incubated with the indicated, purified recombinant GST-Cdh1 or GST-CKIP1 proteins, E1, E2 and ubiquitin as indicated at 30° C for 15 minutes. The ubiquitination reaction products were resolved by SDS-PAGE and probed with the indicated antibodies. (see also Figure S1)
Figure 2. The E3 ligase activity of Smurf1 is augmented by Cdh1
A. Immunoblot (IB) analysis of whole cell lysates (WCL) derived from 293T cells transfected with HA-Smad1 together with Myc-tagged Smurf1 in the presence of increasing amounts of Myc-Cdh1 or Flag-CKIP1. Myc-Cdc20 was included as a negative control. B. Immunoblot (IB) analysis of whole cell lysates (WCL) derived from 293T cells transfected with HA-MEKK2 together with Flag-tagged Smurf1 in the presence of increasing amounts of Myc-Cdh1 or Flag-CKIP1. C. HeLa cells were infected with a shSmurf1 lentiviral vector (with shGFP vector as a negative control) for 24 hours and then selected with 1μg/ml puromycin for 72 hours to eliminate the non-infected cells. The resulting cell lines were transfected with HA-MEKK2 in the presence of increasing amounts of Myc-Cdh1. 40 hours post-transfection, whole cell lysates (WCL) were collected to probe with the indicated antibodies. D. Purified recombinant GST-Cdh1 augments the E3 ligase activity of Smurf1 to ubiquitinate Smad1 in vitro. Bacterially expressed and purified His-Smad1 and GST-Smurf1 were incubated with the indicated, purified recombinant GST-Cdh1 or GST-CKIP1 proteins, purified E1, E2 and ubiquitin as indicated at 30° C for 60 minutes. The ubiquitination reaction products were stopped by the addition of 8 M urea followed by His-tag pull-down in the presence of 8 M urea to eliminate any possible contamination from Smad1-associated proteins. The reactions were then resolved by SDS-PAGE and probed with the indicated antibodies. (see also Figure S2)
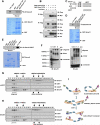
Figure 3. Smurf1 forms dimers in vivo
A. Autoradiography of 35S-labelled Smurf1 bound to the indicated GST fusion proteins. B. Immunoblot (IB) analysis of whole cell lysates (WCL) and immunoprecipitates (IP) derived from 293T cells transfected with HA-Cdh1 and the indicated Smurf1 constructs. Thirty hours post-transfection, cells were pre-treated with 10 μM MG132 for 10 hours to block the proteasome pathway before harvesting. C. Schematic representation of the various Smurf1 truncation mutants used in these studies. D. Autoradiography of 35S-labelled ΔC2-Smurf1 bound to the indicated GST fusion proteins. E. 293T cells were transfected with the HA-Smurf1 HECT construct and 40 hours post-transfection, whole cell lysates (WCL) were recovered to perform GST pull-down analysis with the indicated GST-fusion proteins. F. Immunoblot (IB) analysis of whole cell lysates (WCL) derived from HCT116 cells that are harvested in 0.5% NP40-containing PBS buffer followed by treatment with 0.01% glutaraldehyde for the indicated time periods. Anti-p53 immunoblot was included as a positive control for cross-linking effects. G-H. Gel filtration experiment to illustrate that Smurf1 forms dimers in vivo. Immunoblot analysis of the indicated fractionations derived from the gel filtration experiment with HCT116-shGFP (G) or HCT116-shCdh1 (H) whole cell lysates harvested in EBC buffer. Prior to running cell lysates, the molecular weight resolution of the column was first estimated by running native molecular weight markers (Thyroglobulin ~669KD, Ferritin ~440KD, Aldolase ~158KD, Conalbumin ~75KD and Ovalbumin ~44KD) and determining their retention times on coomassie-stained SDS-PAGE protein gels. I. Proposed model of how the Smurf1 E3 ligase activity is activated by Cdh1 via disruption of Smurf1 homodimer-mediated auto-inhibition. (see also Figure S3)
Figure 4. Cdh1 activates Smurf1 through impairing Smurf1 homodimer-mediated auto-inhibition
A. Immunoblot (IB) analysis of whole cell lysates (WCL) and immunoprecipitates (IP) derived from 293T cells transfected with Flag-Smurf1, Myc-Smurf1 and the indicated HA-tagged Cdh1, CKIP1, Smad1 or Smad7 constructs. Thirty hours post-transfection, cells were pre-treated with 10 μM MG132 for 10 hours to block the proteasome pathway before harvesting. B. Autoradiography of 35S-labelled Smurf1 bound to the indicated GST fusion proteins in the presence of increasing amounts of 35S-labelled Cdh1. C. Autoradiography of 35S-labelled Cdh1 bound to the indicated GST fusion proteins. D. Deletion of the C2 domain augments Smurf1 auto-ubiquitination in vitro. Various bacterially expressed and purified GST-Smurf1 proteins were incubated with purified E1, E2 and ubiquitin as indicated at 30° C for 15 minutes. The ubiquitination reaction products were resolved by SDS-PAGE and probed with the indicated antibodies. E. Deletion of the C2 domain augments the E3 ligase activity of Smurf1 towards ubiquitinating RhoA in vitro. Bacterially expressed and purified His-RhoA and the indicated GST-Smurf1 proteins were incubated with purified E1, E2 and ubiquitin as indicated at 30° C for 60 minutes. The ubiquitination reaction products were stopped by the addition of 8 M urea and subsequent His-tag pull-down was performed in the presence of 8 M urea to eliminate any possible contamination from RhoA-associated proteins before being resolved by SDS-PAGE and probed with the indicated antibodies. (see also Figure S4)
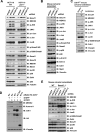
Figure 5. Depletion of Cdh1 leads to inactivation of Smurf1 and subsequent activation of Smurf1 downstream MEKK2 signaling pathway
A. Immunoblot analysis of HCT116-WT or HCT116-_p53_-/- cells infected with the indicated lentiviral shRNA constructs. The infected cells were selected with 1 μg/ml puromycin for 72 hours to eliminate the non-infected cells before harvesting for immunoblot analysis. B. Immunoblot analysis of primary mouse calvarial osteoblast cells infected with the indicated lentiviral shRNA constructs. The infected cells were selected with 1 μg/ml puromycin for 72 hours to eliminate the non-infected cells and the resulting cells were cultured for 6 days under osteoblast differentiation media before harvesting for immunoblot analysis. C. Immunoblot analysis of primary murine _Cdh1_F/F calvarial osteoblast cells, which were derived from neonatal _Cdh1_F/F mice, infected with a lentivirus encoding Cre to deplete endogenous Cdh1 (empty vector infected cells were used as negative controls). D. Immunoblot (IB) analysis of T98G cells infected with the indicated lentiviral shRNA constructs together with the indicated pBabe-Hygro-Cdh1-expression vector. The infected cells were selected with 1μg/ml puromycin and 200 μg/ml hygromycin for 72 hours to eliminate the non-infected cells before harvesting for immunoblot analysis. E. Immunoblot analysis of immortalized WT or _Smurf1_-/- mouse calvarial osteoblast cells infected with the indicated lentiviral shRNA constructs. The infected cells were selected with 1 μg/ml puromycin for 72 hours to eliminate the non-infected cells before harvesting for immunoblot analysis. (see also Figure S5)
Figure 6. Depletion of Cdh1 leads to inactivation of Smurf1 and augmentation of the MEKK2 signaling pathway to induce osteoblast differentiation
A-C. Depletion of Cdh1 in primary mouse calvarial osteoblasts promotes osteoblast differentiation. Primary mouse calvarial osteoblast cells were derived from neonatal mice and infected with the indicated lentiviral shRNA constructs. The infected cells were selected with 1 μg/ml puromycin for 72 hours to eliminate non-infected cells and the efficiency of Cdh1 depletion was examined by anti-Cdh1 immunoblot analysis in (A). Afterwards, the generated cells were incubated in osteoblast differentiation medium for 21 days before the Fast Blue staining for ALP activity (shown as blue staining) and von Kossa staining for minerilization (shown as black staining) in (B). Additionally, the Cdh1 knockdown efficiency and the expression of various characteristic osteoblast markers were determined by real-time PCR analysis in (C). Three sets of independent experiments were performed to generate each data point in (C) and the error bars represent means ± SD. D-E. The osteoblast differentiation is also increased in Cre-encoding lentivirus-induced Cdh1 depletion in primary _Cdh1_F/F calvarial osteoblast cells (empty vector (EV) lentivirus was used as a negative control). The infected cells were selected with 1 μg/ml puromycin for 72 hours to eliminate non-infected cells, the resulting cells were then incubated in osteoblast differentiation medium for 21 days before Fast Blue staining and von Kossa staining in (D). Cre-encoding lentivirus-induced Cdh1 depletion and the expression of various characteristic osteoblast markers were monitored by real-time PCR analysis in (E). Three sets of independent experiments were performed to generate each data point in (E) and the error bars represent means ± SD. F. Ectopic expression of WT-Cdh1 and ΔC-box-Cdh1, but not N174-Cdh1 could suppress the increased osteoblast differentiation phenotype in Cdh1-depleted primary calvarial osteoblast cells. Primary calvarial osteoblast cells derived from _Cdh1_F/F mice were co-infected with Purolentivirus encoding Cre (or a lentiviral vector encoding EGFP as a negative control) and the indicated Hygro-lentiviral vector encoding WT-Cdh1 and various Cdh1 mutants. The infected cells were selected with 1 μg/ml puromycin and 200 μg/ml hygromycin for 96 hours to eliminate non-infected cells. Afterwards, the generated cells were incubated in osteoblast differentiation medium for 21 days before von Kossa staining. (see also Figure S6)
Comment in
- Cdh1 is a HECT of an activator.
Jia L, Yu H. Jia L, et al. Mol Cell. 2011 Dec 9;44(5):681-3. doi: 10.1016/j.molcel.2011.11.012. Mol Cell. 2011. PMID: 22152470
Similar articles
- Non-mitotic functions of the Anaphase-Promoting Complex.
Eguren M, Manchado E, Malumbres M. Eguren M, et al. Semin Cell Dev Biol. 2011 Aug;22(6):572-8. doi: 10.1016/j.semcdb.2011.03.010. Epub 2011 Mar 23. Semin Cell Dev Biol. 2011. PMID: 21439391 Review. - Expression of the CDH1-associated form of the anaphase-promoting complex in postmitotic neurons.
Gieffers C, Peters BH, Kramer ER, Dotti CG, Peters JM. Gieffers C, et al. Proc Natl Acad Sci U S A. 1999 Sep 28;96(20):11317-22. doi: 10.1073/pnas.96.20.11317. Proc Natl Acad Sci U S A. 1999. PMID: 10500174 Free PMC article. - APC/C Cdh1 targets aurora kinase to control reorganization of the mitotic spindle at anaphase.
Floyd S, Pines J, Lindon C. Floyd S, et al. Curr Biol. 2008 Nov 11;18(21):1649-58. doi: 10.1016/j.cub.2008.09.058. Epub 2008 Oct 30. Curr Biol. 2008. PMID: 18976910 - The E3 ligase Cdh1-anaphase promoting complex operates upstream of the E3 ligase Smurf1 in the control of axon growth.
Kannan M, Lee SJ, Schwedhelm-Domeyer N, Stegmüller J. Kannan M, et al. Development. 2012 Oct;139(19):3600-12. doi: 10.1242/dev.081786. Development. 2012. PMID: 22949615 - The emerging role of APC/CCdh1 in development.
Hu D, Qiao X, Wu G, Wan Y. Hu D, et al. Semin Cell Dev Biol. 2011 Aug;22(6):579-85. doi: 10.1016/j.semcdb.2011.03.012. Epub 2011 Apr 7. Semin Cell Dev Biol. 2011. PMID: 21497201 Free PMC article. Review.
Cited by
- Cadherin-mediated cell-cell adhesion and signaling in the skeleton.
Marie PJ, Haÿ E, Modrowski D, Revollo L, Mbalaviele G, Civitelli R. Marie PJ, et al. Calcif Tissue Int. 2014 Jan;94(1):46-54. doi: 10.1007/s00223-013-9733-7. Epub 2013 May 9. Calcif Tissue Int. 2014. PMID: 23657489 Free PMC article. Review. - The protective role of MC1R in chromosome stability and centromeric integrity in melanocytes.
Li X, Mao W, Chen J, Goding CR, Cui R, Xu ZX, Miao X. Li X, et al. Cell Death Discov. 2021 May 18;7(1):111. doi: 10.1038/s41420-021-00499-9. Cell Death Discov. 2021. PMID: 34001865 Free PMC article. - Cdh1-APC Regulates Protein Synthesis and Stress Granules in Neurons through an FMRP-Dependent Mechanism.
Valdez-Sinon AN, Lai A, Shi L, Lancaster CL, Gokhale A, Faundez V, Bassell GJ. Valdez-Sinon AN, et al. iScience. 2020 May 22;23(5):101132. doi: 10.1016/j.isci.2020.101132. Epub 2020 May 1. iScience. 2020. PMID: 32434143 Free PMC article. - Deletion of a core APC/C component reveals APC/C function in regulating neuronal USP1 levels and morphology.
Day JL, Tirard M, Brose N. Day JL, et al. Front Mol Neurosci. 2024 Jun 12;17:1352782. doi: 10.3389/fnmol.2024.1352782. eCollection 2024. Front Mol Neurosci. 2024. PMID: 38932933 Free PMC article. - Ubiquitylation of active Rac1 by the E3 ubiquitin-ligase HACE1.
Mettouchi A, Lemichez E. Mettouchi A, et al. Small GTPases. 2012 Apr-Jun;3(2):102-6. doi: 10.4161/sgtp.19221. Small GTPases. 2012. PMID: 22790197 Free PMC article. Review.
References
- Binne UK, Classon MK, Dick FA, Wei W, Rape M, Kaelin WG, Jr., Naar AM, Dyson NJ. Retinoblastoma protein and anaphase-promoting complex physically interact and functionally cooperate during cell-cycle exit. Nat Cell Biol. 2007;9:225–232. - PubMed
- Burton JL, Tsakraklides V, Solomon MJ. Assembly of an APC-Cdh1-substrate complex is stimulated by engagement of a destruction box. Mol Cell. 2005;18:533–542. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HD055601-04/HD/NICHD NIH HHS/United States
- K01 AG041218/AG/NIA NIH HHS/United States
- R01 GM089763-03/GM/NIGMS NIH HHS/United States
- R01 GM089763-04/GM/NIGMS NIH HHS/United States
- GM089763/GM/NIGMS NIH HHS/United States
- HD55601/HD/NICHD NIH HHS/United States
- R01 HD055601-05/HD/NICHD NIH HHS/United States
- R01 HD055601/HD/NICHD NIH HHS/United States
- R01 GM089763/GM/NIGMS NIH HHS/United States
- R01 GM089763-02/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous