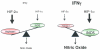HIF1α and HIF2α: sibling rivalry in hypoxic tumour growth and progression - PubMed (original) (raw)
Review
HIF1α and HIF2α: sibling rivalry in hypoxic tumour growth and progression
Brian Keith et al. Nat Rev Cancer. 2011.
Abstract
Hypoxia-inducible factors (HIFs) are broadly expressed in human cancers, and HIF1α and HIF2α were previously suspected to promote tumour progression through largely overlapping functions. However, this relatively simple model has now been challenged in light of recent data from various approaches that reveal unique and sometimes opposing activities of these HIFα isoforms in both normal physiology and disease. These effects are mediated in part through the regulation of unique target genes, as well as through direct and indirect interactions with important oncoproteins and tumour suppressors, including MYC and p53. As HIF inhibitors are currently undergoing clinical evaluation as cancer therapeutics, a more thorough understanding of the unique roles performed by HIF1α and HIF2α in human neoplasia is warranted.
Figures
Figure 1. HIF1α and HIF2α exhibit antagonistic functions in nitric oxide (NO) production
Under low IFNy conditions, HIF2α is more abundant and induces arginase1 expression, resulting in NO production. Under high IFNy conditions, HIF2α is diminished and HIF1α dominates so that iNOS can utilize arginine for NO generation. These physiologically antagonistic functions allow the HIFα subunits to coordinately regulate NO production in a cytokine-induced and transcription-dependent fashion.
Figure 2. HIF1α and HIF2α are post-translationally modified, and differentially regulated by multiple mechanisms
(A) Multiple mechanisms differentially regulate HIF1α and HIF2α at the levels of transcription or mRNA stability (red), mRNA translation (green), and protein stability (blue). In most cases, these regulatory events have opposite effects on HIF1α and HIF2α expression, or appear to be specific for only one HIFα isoform. See text for details. (NE), no effect. (8) Summary of phosphorylations, acetylations, and hydroxylations of the two HIFα subunits by CK1, ARD1, PHDs, FIH, MAPK, SIRT1, PKD1, and ATM. It should be noted that ARD1 acetylates HIF1α, while SIRT1 deacetylates both HIF1α and HIF2α. (C) Sequence alignment of HIF2α residues 301-331 with a similar region of HIF1α; shaded residues are unique to HIF2α and allow the selective phosphorylation of HIF2α T324 by PKD1.
Figure 2. HIF1α and HIF2α are post-translationally modified, and differentially regulated by multiple mechanisms
(A) Multiple mechanisms differentially regulate HIF1α and HIF2α at the levels of transcription or mRNA stability (red), mRNA translation (green), and protein stability (blue). In most cases, these regulatory events have opposite effects on HIF1α and HIF2α expression, or appear to be specific for only one HIFα isoform. See text for details. (NE), no effect. (8) Summary of phosphorylations, acetylations, and hydroxylations of the two HIFα subunits by CK1, ARD1, PHDs, FIH, MAPK, SIRT1, PKD1, and ATM. It should be noted that ARD1 acetylates HIF1α, while SIRT1 deacetylates both HIF1α and HIF2α. (C) Sequence alignment of HIF2α residues 301-331 with a similar region of HIF1α; shaded residues are unique to HIF2α and allow the selective phosphorylation of HIF2α T324 by PKD1.
Figure 2. HIF1α and HIF2α are post-translationally modified, and differentially regulated by multiple mechanisms
(A) Multiple mechanisms differentially regulate HIF1α and HIF2α at the levels of transcription or mRNA stability (red), mRNA translation (green), and protein stability (blue). In most cases, these regulatory events have opposite effects on HIF1α and HIF2α expression, or appear to be specific for only one HIFα isoform. See text for details. (NE), no effect. (8) Summary of phosphorylations, acetylations, and hydroxylations of the two HIFα subunits by CK1, ARD1, PHDs, FIH, MAPK, SIRT1, PKD1, and ATM. It should be noted that ARD1 acetylates HIF1α, while SIRT1 deacetylates both HIF1α and HIF2α. (C) Sequence alignment of HIF2α residues 301-331 with a similar region of HIF1α; shaded residues are unique to HIF2α and allow the selective phosphorylation of HIF2α T324 by PKD1.
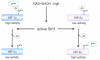
Figure 3. Differential regulation of HIF1α and HIF2α by SIRT1
(A) High levels of NAD+ inactivate SIRT1, resulting in decreased HIF1α transcriptional activity and enhanced HIF2α stimulation of target genes like erythropoietin. (8) Distinct effects of HIF1α and HIF2α on MYC complex formation and promoter occupancy. Hypoxic cells exclusively expressing HIF1α exhibit decreased MYC activity due to diminished association with MAX and SP1, as well as reduced MYC stability. HIF1α also induces MXI1 expression, which inhibits MYC target gene expression (see text for details). Cells expressing HIF2α exhibit increased MYC complex formation and target gene activation, although the mechanisms involved are not fully understood.
Figure 3. Differential regulation of HIF1α and HIF2α by SIRT1
(A) High levels of NAD+ inactivate SIRT1, resulting in decreased HIF1α transcriptional activity and enhanced HIF2α stimulation of target genes like erythropoietin. (8) Distinct effects of HIF1α and HIF2α on MYC complex formation and promoter occupancy. Hypoxic cells exclusively expressing HIF1α exhibit decreased MYC activity due to diminished association with MAX and SP1, as well as reduced MYC stability. HIF1α also induces MXI1 expression, which inhibits MYC target gene expression (see text for details). Cells expressing HIF2α exhibit increased MYC complex formation and target gene activation, although the mechanisms involved are not fully understood.
Similar articles
- Drug resistance and cancer stem cells: the shared but distinct roles of hypoxia-inducible factors HIF1α and HIF2α.
Schöning JP, Monteiro M, Gu W. Schöning JP, et al. Clin Exp Pharmacol Physiol. 2017 Feb;44(2):153-161. doi: 10.1111/1440-1681.12693. Clin Exp Pharmacol Physiol. 2017. PMID: 27809360 Review. - A macrophage-dominant PI3K isoform controls hypoxia-induced HIF1α and HIF2α stability and tumor growth, angiogenesis, and metastasis.
Joshi S, Singh AR, Zulcic M, Durden DL. Joshi S, et al. Mol Cancer Res. 2014 Oct;12(10):1520-31. doi: 10.1158/1541-7786.MCR-13-0682. Epub 2014 Aug 7. Mol Cancer Res. 2014. PMID: 25103499 - Opposing effects of HIF1α and HIF2α on chromaffin cell phenotypic features and tumor cell proliferation: Insights from MYC-associated factor X.
Qin N, de Cubas AA, Garcia-Martin R, Richter S, Peitzsch M, Menschikowski M, Lenders JW, Timmers HJ, Mannelli M, Opocher G, Economopoulou M, Siegert G, Chavakis T, Pacak K, Robledo M, Eisenhofer G. Qin N, et al. Int J Cancer. 2014 Nov 1;135(9):2054-64. doi: 10.1002/ijc.28868. Epub 2014 Apr 7. Int J Cancer. 2014. PMID: 24676840 - The HIF1α/HIF2α-miR210-3p network regulates glioblastoma cell proliferation, dedifferentiation and chemoresistance through EGF under hypoxic conditions.
Wang P, Yan Q, Liao B, Zhao L, Xiong S, Wang J, Zou D, Pan J, Wu L, Deng Y, Wu N, Gong S. Wang P, et al. Cell Death Dis. 2020 Nov 18;11(11):992. doi: 10.1038/s41419-020-03150-0. Cell Death Dis. 2020. PMID: 33208727 Free PMC article. - Hypoxia and hypoxia inducible factors in cancer stem cell maintenance.
Li Z, Rich JN. Li Z, et al. Curr Top Microbiol Immunol. 2010;345:21-30. doi: 10.1007/82_2010_75. Curr Top Microbiol Immunol. 2010. PMID: 20582533 Review.
Cited by
- Elucidation of the Role of SHMT2 in L-Serine Homeostasis in Hypoxic Hepa1-6 Cells.
Zhang S, He R, Zhang M, Zhang J, Wu M, Zhang G, Jiang T. Zhang S, et al. Int J Mol Sci. 2024 Nov 2;25(21):11786. doi: 10.3390/ijms252111786. Int J Mol Sci. 2024. PMID: 39519335 Free PMC article. - ATF4/NUPR1 axis promotes cancer cell survival and mediates immunosuppression in clear cell renal cell carcinoma.
Lu Y, Chen W, Xuan Y, Li X, Wu S, Wang H, Guo T, Wang C, Tian S, Li H, Lai D, Zhao W, Huang X, Zhao X, Wang B, Zhang X, Li H, Huang Y, Ma X. Lu Y, et al. Discov Oncol. 2024 Oct 31;15(1):607. doi: 10.1007/s12672-024-01485-0. Discov Oncol. 2024. PMID: 39480570 Free PMC article. - Glucose deprivation impairs hypoxia-inducible factor-1α synthesis.
Hubert M, Stuart S, Ohh M. Hubert M, et al. Discov Oncol. 2024 Oct 28;15(1):595. doi: 10.1007/s12672-024-01484-1. Discov Oncol. 2024. PMID: 39466364 Free PMC article. - Pilot Study on the Effect of Patient Condition and Clinical Parameters on Hypoxia-Induced Factor Expression: HIF1A, EPAS1 and HIF3A in Human Colostrum Cells.
Zarychta J, Kowalczyk A, Słowik K, Przywara D, Petniak A, Kondracka A, Wójtowicz-Marzec M, Słyk-Gulewska P, Kwaśniewska A, Kocki J, Gil-Kulik P. Zarychta J, et al. Int J Mol Sci. 2024 Oct 14;25(20):11042. doi: 10.3390/ijms252011042. Int J Mol Sci. 2024. PMID: 39456823 Free PMC article. - Hypoxia sensing in resident cardiac macrophages regulates monocyte fate specification following ischemic heart injury.
Kadyrov FF, Koenig AL, Amrute JM, Dun H, Li W, Weinheimer CJ, Nigro JM, Kovacs A, Bredemeyer AL, Yang S, Das S, Penna VR, Parvathaneni A, Lai L, Hartmann N, Kopecky BJ, Kreisel D, Lavine KJ. Kadyrov FF, et al. Nat Cardiovasc Res. 2024 Nov;3(11):1337-1355. doi: 10.1038/s44161-024-00553-6. Epub 2024 Oct 21. Nat Cardiovasc Res. 2024. PMID: 39433910
References
- Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. - PubMed
- Kaelin WG, Jr., Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous