Whole genome sequencing of matched primary and metastatic acral melanomas - PubMed (original) (raw)
Case Reports
. 2012 Feb;22(2):196-207.
doi: 10.1101/gr.125591.111. Epub 2011 Dec 19.
Simon J Furney, Maryou B Lambros, Costas Mitsopoulos, Iwanka Kozarewa, Felipe C Geyer, Alan Mackay, Jarle Hakas, Marketa Zvelebil, Christopher J Lord, Alan Ashworth, Meirion Thomas, Gordon Stamp, James Larkin, Jorge S Reis-Filho, Richard Marais
Affiliations
- PMID: 22183965
- PMCID: PMC3266028
- DOI: 10.1101/gr.125591.111
Case Reports
Whole genome sequencing of matched primary and metastatic acral melanomas
Samra Turajlic et al. Genome Res. 2012 Feb.
Abstract
Next generation sequencing has enabled systematic discovery of mutational spectra in cancer samples. Here, we used whole genome sequencing to characterize somatic mutations and structural variation in a primary acral melanoma and its lymph node metastasis. Our data show that the somatic mutational rates in this acral melanoma sample pair were more comparable to the rates reported in cancer genomes not associated with mutagenic exposure than in the genome of a melanoma cell line or the transcriptome of melanoma short-term cultures. Despite the perception that acral skin is sun-protected, the dominant mutational signature in these samples is compatible with damage due to ultraviolet light exposure. A nonsense mutation in ERCC5 discovered in both the primary and metastatic tumors could also have contributed to the mutational signature through accumulation of unrepaired dipyrimidine lesions. However, evidence of transcription-coupled repair was suggested by the lower mutational rate in the transcribed regions and expressed genes. The primary and the metastasis are highly similar at the level of global gene copy number alterations, loss of heterozygosity and single nucleotide variation (SNV). Furthermore, the majority of the SNVs in the primary tumor were propagated in the metastasis and one nonsynonymous coding SNV and one splice site mutation appeared to arise de novo in the metastatic lesion.
Figures
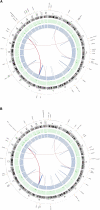
Figure 1.
Summary of lesions found in acral melanoma. Shown are primary and metastasis Circos plots (Krzywinski et al. 2009) of somatic mutations in the primary (A) and metastatic (B) tumors. The outer circle contains whole genome high-confidence SNVs (black dots) and nonsynonymous SNVs (orange dots; annotated with HGNC/Ensembl gene symbols). Copy number alterations are shown in the inner two plots (green circle shows gains and blue shows losses). Validated structural variations are depicted as links in the interior of the plot.
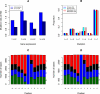
Figure 2.
Somatic SNVs analysis in acral melanoma samples. (A) Somatic SNV mutation rates of all SNVs and C>T/G>A transitions for expressed genes in the primary (P-EXP) and metastatic tumors (M-EXP) and nonexpressed genes (P-NON and M-NON). (B) Proportion of somatic SNVs by class in the primary and metastatic tumors for the whole genome (WG) and coding regions (CDS). (C) Frequency of bases ±5 bp of whole genome C>T/G>A transitions in the primary tumors. (D) Frequency of bases ±5 bp of whole genome C>T/G>A transitions in the metastatic tumors.
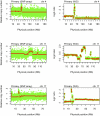
Figure 3.
Copy number alterations in the primary tumor. SNP array plots showing somatic copy number alteration (CNA) in the primary tumor detected by genome-wide SNP arrays (left column) and whole-genome (WG) sequencing normalized read-depth per 10 kb (right column) for chromosomes 4, 11, and 17.
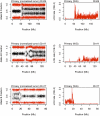
Figure 4.
Loss of heterozygosity in the primary tumor. SNP array plots showing loss of heterozygosity (LOH) in the primary tumor as detected by allele B frequency from SNP array (left column) and whole-genome sequencing LOH rate in 100-kb windows (right column) for chromosomes 4, 11, and 17.
Similar articles
- The mutational burden of acral melanoma revealed by whole-genome sequencing and comparative analysis.
Furney SJ, Turajlic S, Stamp G, Thomas JM, Hayes A, Strauss D, Gavrielides M, Xing W, Gore M, Larkin J, Marais R. Furney SJ, et al. Pigment Cell Melanoma Res. 2014 Sep;27(5):835-8. doi: 10.1111/pcmr.12279. Epub 2014 Jun 30. Pigment Cell Melanoma Res. 2014. PMID: 24913711 - Genome sequencing of mucosal melanomas reveals that they are driven by distinct mechanisms from cutaneous melanoma.
Furney SJ, Turajlic S, Stamp G, Nohadani M, Carlisle A, Thomas JM, Hayes A, Strauss D, Gore M, van den Oord J, Larkin J, Marais R. Furney SJ, et al. J Pathol. 2013 Jul;230(3):261-9. doi: 10.1002/path.4204. J Pathol. 2013. PMID: 23620124 - Mutational signatures of de-differentiation in functional non-coding regions of melanoma genomes.
Parker SC, Gartner J, Cardenas-Navia I, Wei X, Ozel Abaan H, Ajay SS, Hansen NF, Song L, Bhanot UK, Killian JK, Gindin Y, Walker RL, Meltzer PS, Mullikin JC, Furey TS, Crawford GE, Rosenberg SA, Samuels Y, Margulies EH. Parker SC, et al. PLoS Genet. 2012;8(8):e1002871. doi: 10.1371/journal.pgen.1002871. Epub 2012 Aug 9. PLoS Genet. 2012. PMID: 22912592 Free PMC article. - The genomic landscape of cutaneous melanoma.
Zhang T, Dutton-Regester K, Brown KM, Hayward NK. Zhang T, et al. Pigment Cell Melanoma Res. 2016 May;29(3):266-83. doi: 10.1111/pcmr.12459. Epub 2016 Mar 4. Pigment Cell Melanoma Res. 2016. PMID: 26833684 Review. - Molecular Pathways in Melanomagenesis: What We Learned from Next-Generation Sequencing Approaches.
Palmieri G, Colombino M, Casula M, Manca A, Mandalà M, Cossu A; Italian Melanoma Intergroup (IMI). Palmieri G, et al. Curr Oncol Rep. 2018 Sep 14;20(11):86. doi: 10.1007/s11912-018-0733-7. Curr Oncol Rep. 2018. PMID: 30218391 Free PMC article. Review.
Cited by
- Future paradigms for precision oncology.
Klement GL, Arkun K, Valik D, Roffidal T, Hashemi A, Klement C, Carmassi P, Rietman E, Slaby O, Mazanek P, Mudry P, Kovacs G, Kiss C, Norga K, Konstantinov D, André N, Slavc I, van Den Berg H, Kolenova A, Kren L, Tuma J, Skotakova J, Sterba J. Klement GL, et al. Oncotarget. 2016 Jul 19;7(29):46813-46831. doi: 10.18632/oncotarget.9488. Oncotarget. 2016. PMID: 27223079 Free PMC article. Review. - From melanocytes to melanomas.
Shain AH, Bastian BC. Shain AH, et al. Nat Rev Cancer. 2016 Jun;16(6):345-58. doi: 10.1038/nrc.2016.37. Epub 2016 Apr 29. Nat Rev Cancer. 2016. PMID: 27125352 Review. - Implications of genetic heterogeneity in cancer.
Schmitt MW, Prindle MJ, Loeb LA. Schmitt MW, et al. Ann N Y Acad Sci. 2012 Sep;1267:110-6. doi: 10.1111/j.1749-6632.2012.06590.x. Ann N Y Acad Sci. 2012. PMID: 22954224 Free PMC article. - Evolution and dynamics of pancreatic cancer progression.
Yachida S, Iacobuzio-Donahue CA. Yachida S, et al. Oncogene. 2013 Nov 7;32(45):5253-60. doi: 10.1038/onc.2013.29. Epub 2013 Feb 18. Oncogene. 2013. PMID: 23416985 Free PMC article. Review. - Case report of a Li-Fraumeni syndrome-like phenotype with a de novo mutation in CHEK2.
Zhuang X, Li Y, Cao H, Wang T, Chen J, Liu J, Lin L, Ye R, Li X, Liu S, Li W, Lv Y, Zhang J, He C, Xu X, Wang Z, Huang C, Liu X, Wang L. Zhuang X, et al. Medicine (Baltimore). 2016 Jul;95(29):e4251. doi: 10.1097/MD.0000000000004251. Medicine (Baltimore). 2016. PMID: 27442652 Free PMC article.
References
- Albert SM 2010. Neurodegenerative disease and cancer: A critical role for melanoma? Neuroepidemiology 35: 305–306 - PubMed
- Aung CS, Hill MM, Bastiani M, Parton RG, Parat MO 2011. PTRF-cavin-1 expression decreases the migration of PC3 prostate cancer cells: Role of matrix metalloprotease 9. Eur J Cell Biol 90: 136–142 - PubMed
- Bastian BC, Kashani-Sabet M, Hamm H, Godfrey T, Moore DH 2nd, Brocker EB, LeBoit PE, Pinkel D 2000. Gene amplifications characterize acral melanoma and permit the detection of occult tumor cells in the surrounding skin. Cancer Res 60: 1968–1973 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases