Ubiquitylation of the nuclear pore complex controls nuclear migration during mitosis in S. cerevisiae - PubMed (original) (raw)
Ubiquitylation of the nuclear pore complex controls nuclear migration during mitosis in S. cerevisiae
Akira Hayakawa et al. J Cell Biol. 2012.
Abstract
Nuclear pore complexes (NPCs) correspond to large protein transport complexes responsible for selective nucleocytoplasmic exchange. Although research has revealed much about the molecular architecture and roles of the NPC subcomplexes, little is known about the regulation of NPC functions by posttranslational modifications. We used a systematic approach to show that more than half of NPC proteins were conjugated to ubiquitin. In particular, Nup159, a nucleoporin exclusively located on the cytoplasmic side of the NPC, was monoubiquitylated by the Cdc34/SCF (Skp1-Cdc53-F-box E3 ligase) enzymes. Preventing this modification had no consequences on nuclear transport or NPC organization but strongly affected the ability of Nup159 to target the dynein light chain to the NPC. This led to defects in nuclear segregation at the onset of mitosis. Thus, defining ubiquitylation of the yeast NPC highlights yet-unexplored functions of this essential organelle in cell division.
Figures
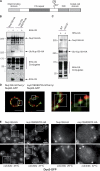
Figure 1.
Ubiquitylation of nucleoporins. Ni-purified 6His-ubiquitin (Ub)–conjugated forms of genomically HA-tagged Nups were extracted from cells transformed (+) or not transformed (−) with a plasmid encoding 6His-ubiquitin under control of the CUP1 promoter. Cell lysates and Ni-purified material were examined by Western blotting with an anti-HA antibody. Ubiquitin expression and efficiency of purification were controlled using an anti-6His antibody (not depicted). Localization of Nups as previously determined (Rout et al., 2000) is indicated on the right (N, nuclear side of the NPC; C, cytoplasmic side of the NPC). (A) Analysis of ubiquitin-conjugated forms of genomically HA-tagged FG-Nups. (B) Analysis of ubiquitin-conjugated forms of genomically HA-tagged transmembrane Nups. (C) Summary of the ubiquitylation status of Nups. Molecular masses are given in kilodaltons.
Figure 2.
Ubiquitylation of Nup159 and its role in Dyn2 localization at the NPC. (A) Schematic representation of Nup159 molecular organization (DID, dynein-interacting domain). (B) Ni-purified 6His-ubiquitin (Ub)–conjugated forms of Nup159-HA were extracted from cells transformed (+) or not transformed (−) with a plasmid encoding 6His-ubiquitin under control of the CUP1 promoter. Cell lysates (top) and Ni-purified material (middle) were examined by Western blotting with an anti-HA antibody. A portion of the Ni-purified material also was examined by Western blotting on a separate membrane using an anti-6His antibody to show purification efficiency (bottom). (B and C) Analysis of ubiquitin-conjugated forms of Nup159-HA was performed in wt (Nup159-HA) and _nup159K897R_-HA cells (B) and in wt and cdc34ts strains (C). The white line indicates that intervening lanes have been spliced out. Molecular markers are given in kilodaltons. (D) Localization of genomically tagged Nup60-GFP and Nup159-mCherry (left) or Nup60-GFP and _nup159K897R_-mCherry was monitored by SIM. Centroid distances of ∼90 ± 20 nm, along the line axis indicated in the magnified areas, were estimated between the two fluorescent signals at the NPC (n = 20 for each cell types). Bars, 1 µm. (E) Localization of genomically tagged Dyn2-GFP was analyzed in Nup159-HA and nup159K897R_-HA cells as well as in c_dc34ts cells at 23°C and after a shift to 37°C. NPC localization corresponds to the rimlike staining. The magnified areas (boxes) are shown on the right. Bars, 5 µm.
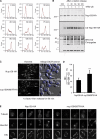
Figure 3.
Ubiquitylation of Nup159 facilitates nuclear migration to the daughter cells during mitosis. (A) Ubiquitylation of Nup159 does not affect cell cycle progression. Nup159-HA (black) and _nup159K897R_-HA (red) cells were synchronized with α-factor and then released for the indicated period of time before fixation and staining with SYTOX green and analyzed by flow cytometry. Numbers on the bottoms show fluorescence intensity (arbitrary units). (B) Ubiquitylation of Nup159 is not cell cycle dependent. Ni-purified 6His-ubiquitin (Ub)–conjugated forms of Nup159-HA were extracted from unsynchronized (O/N, overnight) cells or from cells treated with α-factor for 3 h before release for indicated periods of time. Cell lysates (top) and Ni-purified material (middle) were examined by Western blotting with an anti-HA antibody. (bottom) Ubiquitin expression and efficiency of purification were controlled using an anti-6His antibody. Molecular markers are given in kilodaltons. Numbers at the top are in minutes. (C) Cells were treated with α-factor for 3 h, released for 90 min, fixed, and stained with Hoechst before epifluorescence and interferential contrast analysis. Dotted lines show the yeast cell contours. Arrows indicate daughter cells. (D) Segregation of nuclei observed in A was analyzed in >100 wt and mutant cells per experiment, and results were obtained from five independent experiments. Error bars show standard deviations. ***, P < 0.05. (E) Localization of microtubules was analyzed by fluorescence microscopy in Nup159-HA and _nup159K897R_-HA cells using Tub1-GFP. DIC, differential interference contrast. Bars, 5 µm.
Figure 4.
Ubiquitylation of Nup159 is in the dynein pathway not the Kar9 pathway. (A) Serial dilutions of the corresponding strains were spotted onto YPD plates and grown at the indicated temperature. (B) _kar9Δ_Nup159-HA and _kar9Δnup159K897R_-HA cells grown at 30°C were fixed and stained with Hoechst before epifluorescence and interferential contrast analysis. Arrows indicate some defecting cells. DIC, differential interference contrast. Bar, 5 µm.
Comment in
- A monoubiquitylation pore anchor.
Schuldt A. Schuldt A. Nat Rev Mol Cell Biol. 2012 Jan 18;13(2):66. doi: 10.1038/nrm3276. Nat Rev Mol Cell Biol. 2012. PMID: 22251900 No abstract available.
Similar articles
- Molecular basis for the functional interaction of dynein light chain with the nuclear-pore complex.
Stelter P, Kunze R, Flemming D, Höpfner D, Diepholz M, Philippsen P, Böttcher B, Hurt E. Stelter P, et al. Nat Cell Biol. 2007 Jul;9(7):788-96. doi: 10.1038/ncb1604. Epub 2007 Jun 3. Nat Cell Biol. 2007. PMID: 17546040 - Cdc34 C-terminal tail phosphorylation regulates Skp1/cullin/F-box (SCF)-mediated ubiquitination and cell cycle progression.
Sadowski M, Mawson A, Baker R, Sarcevic B. Sadowski M, et al. Biochem J. 2007 Aug 1;405(3):569-81. doi: 10.1042/BJ20061812. Biochem J. 2007. PMID: 17461777 Free PMC article. - NPC-phagy: selective autophagy of the nuclear pore complexes.
Yin Z, Klionsky DJ. Yin Z, et al. Autophagy. 2020 Oct;16(10):1735-1736. doi: 10.1080/15548627.2020.1798199. Epub 2020 Jul 27. Autophagy. 2020. PMID: 32713250 Free PMC article. - [Nuclear pores: from yeast to higher eukaryotes].
Doye V. Doye V. J Soc Biol. 2002;196(4):349-54. J Soc Biol. 2002. PMID: 12645306 Review. French. - Structure and Assembly of the Nuclear Pore Complex.
Hampoelz B, Andres-Pons A, Kastritis P, Beck M. Hampoelz B, et al. Annu Rev Biophys. 2019 May 6;48:515-536. doi: 10.1146/annurev-biophys-052118-115308. Epub 2019 Apr 3. Annu Rev Biophys. 2019. PMID: 30943044 Review.
Cited by
- Cytoplasmic nucleoporin assemblage: the cellular artwork in physiology and disease.
Lin J, Sumara I. Lin J, et al. Nucleus. 2024 Dec;15(1):2387534. doi: 10.1080/19491034.2024.2387534. Epub 2024 Aug 12. Nucleus. 2024. PMID: 39135336 Free PMC article. Review. - The yeast Ty1 retrotransposon requires components of the nuclear pore complex for transcription and genomic integration.
Manhas S, Ma L, Measday V. Manhas S, et al. Nucleic Acids Res. 2018 Apr 20;46(7):3552-3578. doi: 10.1093/nar/gky109. Nucleic Acids Res. 2018. PMID: 29514267 Free PMC article. - Combining Spinach-tagged RNA and gene localization to image gene expression in live yeast.
Guet D, Burns LT, Maji S, Boulanger J, Hersen P, Wente SR, Salamero J, Dargemont C. Guet D, et al. Nat Commun. 2015 Nov 19;6:8882. doi: 10.1038/ncomms9882. Nat Commun. 2015. PMID: 26582123 Free PMC article. - Uncovering nuclear pore complexity with innovation.
Adams RL, Wente SR. Adams RL, et al. Cell. 2013 Mar 14;152(6):1218-21. doi: 10.1016/j.cell.2013.02.042. Cell. 2013. PMID: 23498931 Free PMC article. Review. - SUMOylation of the nuclear pore complex basket is involved in sensing cellular stresses.
Folz H, Niño CA, Taranum S, Caesar S, Latta L, Waharte F, Salamero J, Schlenstedt G, Dargemont C. Folz H, et al. J Cell Sci. 2019 Apr 3;132(7):jcs224279. doi: 10.1242/jcs.224279. J Cell Sci. 2019. PMID: 30837289 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials