CBX7 is a tumor suppressor in mice and humans - PubMed (original) (raw)
. 2012 Feb;122(2):612-23.
doi: 10.1172/JCI58620. Epub 2012 Jan 3.
Antonella Federico, Pierlorenzo Pallante, Adele Abbate, Francesco Esposito, Umberto Malapelle, Romina Sepe, Giuseppe Palma, Giancarlo Troncone, Marzia Scarfò, Claudio Arra, Monica Fedele, Alfredo Fusco
Affiliations
- PMID: 22214847
- PMCID: PMC3266782
- DOI: 10.1172/JCI58620
CBX7 is a tumor suppressor in mice and humans
Floriana Forzati et al. J Clin Invest. 2012 Feb.
Erratum in
- J Clin Invest. 2013 Feb 1;123(2):934
Abstract
The CBX7 gene encodes a polycomb group protein that is known to be downregulated in many types of human cancers, although the role of this protein in carcinogenesis remains unclear. To shed light on this issue, we generated mice null for Cbx7. Mouse embryonic fibroblasts derived from these mice had a higher growth rate and reduced susceptibility to senescence compared with their WT counterparts. This was associated with upregulated expression of multiple cell cycle components, including cyclin E, which is known to play a key role in lung carcinogenesis in humans. Adult Cbx7-KO mice developed liver and lung adenomas and carcinomas. In in vivo and in vitro experiments, we demonstrated that CBX7 bound to the CCNE1 promoter in a complex that included HDAC2 and negatively regulated CCNE1 expression. Finally, we found that the lack of CBX7 protein expression in human lung carcinomas correlated with CCNE1 overexpression. These data suggest that CBX7 is a tumor suppressor and that its loss plays a key role in the pathogenesis of cancer.
Figures
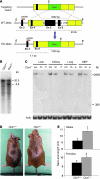
Figure 1. Generation of _Cbx7_-KO mice.
(A) Endogenous WT allele, targeting vector, and resulting KO allele. E, _Eco_RI; Neo, neomycin; Ex, exon. (B) Southern blot of representative Cbx7+/+ and Cbx7+/– ES cell clones. (C) Northern blot analysis of total RNA from kidneys, lungs and MEFs of Cbx7+/+, Cbx7+/–, and Cbx7–/– mice. The lengths of the WT Cbx7 transcript (2,900 bp) and the predicted truncated form after homologous recombination with the _Cbx7_-KO construct (380 bp) are shown. The absence of a 380-bp signal in both Cbx7+/– and Cbx7–/– mice is likely the result of instability of the short transcript. Actb expression analysis of the same blot is shown as control of RNA loaded. (D) Gross appearance of a representative 1-year-old Cbx7–/– mouse and a sex-matched Cbx7+/+ sibling. (E) Naso-anal length of cohorts of 20 mice, males or females, was measured at 12 months of age. Values are mean ± SD. *P < 0.05.
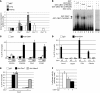
Figure 2. Growth alterations in _Cbx7_-KO MEFs.
(A) MEFs were prepared from Cbx7+/+, Cbx7+/–, and Cbx7–/– embryos at 12.5 dpc. At culture passage 4, they were plated and counted daily for 13 days to extrapolate growth curves. Shown are mean ± SEM of 3 different cell clones (each originating from a different embryo) for each genotype. (B and C) Propidium iodide flow cytometry of asynchronous growing WT and _Cbx7_-KO MEFs. (B) Percent cells in each phase of the cell cycle (mean ± SEM). *P < 0.05. (C) FACS analysis of Cbx7–/– MEFs transiently transfected with different amounts of a Cbx7 expression vector or the backbone vector (BV). (D–I) Light microscopy of representative Cbx7+/+ (D and G), Cbx7+/– (E and H), and Cbx7–/– (F and I) MEF clones stained for β-galactosidase activity at culture passages 4 (P4; D–F) and 7 (G–I). (J) Expression of cell cycle inhibitors Cdkn2a, tp53, and Cdkn1a in representative MEFs from each genotype was determined by qRT-PCR at culture passages 4 and 7. *P < 0.05. Passage 4 Cbx7–/– MEFs transiently transfected with the backbone vector (C) or whose Cbx7 expression had been restored (R) are also shown. (K) Expression of cell cycle and senescence regulators in representative MEFs from each genotype was determined by Western blot at culture passage 7. Normalized protein levels, evaluated by densitometric analysis, are indicated above immunoblots.
Figure 3. CBX7-dependent regulation of Ccne1 expression.
(A) qRT-PCR on MEFs and tissues from WT and _Cbx7_-KO mice, including Cbx7–/– MEFs transiently transfected with the backbone vector (_Cbx7–/–_C) or whose Cbx7 expression had been restored (_Cbx7–/–_R), to detect Ccne1 expression. (B) EMSA performed with a radiolabeled CCNE1 promoter oligonucleotide incubated with 5 and 20 ng of the recombinant GST-CBX7 (lanes 2 and 3), GST-CBX7-CHROMO (lanes 5 and 6), or GST-CBX7-NOCHROMO (lanes 8 and 9) proteins. Where indicated, a 400-fold molar excess of unlabeled CCNE1 promoter oligonucleotide was added (lanes 4, 7, and 10). Further negative controls were obtained by incubating the recombinant proteins with a radiolabeled GAPDH promoter oligonucleotide (lanes 11 and 12). (C–E) Results of ChIP assays. CDKN2A and CDH1 promoters were used as positive controls, whereas GAPDH promoter and nonspecific IgG instead of anti-CBX7 were used as negative controls. (C) ChIP assay on HEK 293 cells transfected with _CBX7_-expressing or empty vector for binding of CBX7 to the CCNE1 promoter. (D) ChIP assay on MEFs for binding of endogenous Cbx7 to the Ccne1 promoter. (E) ChIP assay on MEFs for binding of endogenous Cbx7 and Hdac2 to the Ccne1 promoter. (F) CCNE1 promoter–driven luciferase activity, relative to activation of empty vector–transfected cells, in HEK 293 cells. Where indicated, 1 and 2 μg of CBX7 was cotransfected with the cyclin E–luc plasmid. *P < 0.05; **P < 0.01.
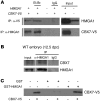
Figure 4. CBX7 interacts with HMGA1b.
(A) HEK 293 cells were transfected with both CBX7-V5 and HMGA1b expression plasmids. Cellular lysates were prepared, and equal amounts of proteins were subjected to IP with anti–CBX7-V5, anti-HMGA1, or nonspecific IgG, as indicated. The immunocomplexes were immunoblotted with reciprocal antibodies. As a positive control, 50 μg of transfected cell lysates were separated on the polyacrylamide gel as input. Et.Br., ethidium bromide. (B) Tissue lysates were prepared from WT embryos, and equal amounts of proteins were subjected to IP with anti-HMGA1 antibodies or nonspecific IgG. The immunocomplexes were immunoblotted with anti-CBX7 antibodies, for detection of the co-IP, or with anti-HMGA1 antibodies, for the IP control. As a positive control, 100 μg tissue lysates were used as input. Lanes were run on the same gel but were noncontiguous (white lines). (C) GST-HMGA1b or GST proteins immobilized on glutathione beads were used to bind CBX7-V5 from HEK 293 cells. The filter was probed with the anti–CBX7-V5 antibody.
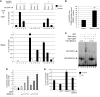
Figure 5. Competition between HMGA1 and CBX7 for binding, regulation, and acetylation of the CCNE1 promoter.
(A) ChIP and re-ChIP assay, revealed by qRT-PCR, on HEK 293 cells transfected with HMGA1b expression vector, HA-tagged CBX7 expression vector, or both to detect the HMGA1/CBX7, CBX7/HDAC2, and HMGA1/HDAC2 interactions on the CCNE1 promoter. GAPDH promoter was also analyzed as a negative control. (B) Percent HDAC2 protein co-IP with HMGA1, as detected in the ChIP and re-ChIP assay in A, comparatively reported in cells transfected with HMGA1b with or without CBX7. **P < 0.01. (C) EMSA, performed as in Figure 3B, using 5 ng GST-CBX7 (lanes 4–8) and 5 and 20 ng His-HMGA1b (lanes 1–3 and 6–8). A 400-fold molar excess of unlabeled probe (lanes 3, 5, and 8) was added as a specific competitor, and 5 ng of a recombinant GST protein (lane 9) was used as a negative control. (D) CCNE1 promoter–driven luciferase activity, expressed relative to activation of empty vector–transfected cells, in HEK 293 cells. Where indicated, 1 μg CBX7 and/or 1, 2, and 5 μg HMGA1b expression vectors were cotransfected with the cyclin E–luc plasmid. (E) ChIP assay, revealed by qRT-PCR, on HEK 293 cells transfected with CBX7 expression vector, HMGA1b expression vector, both, or empty vector for binding of acetylated histone H4 (AcH4) to the CCNE1 promoter. Equal amounts of proteins were subjected to IP with anti-AcH4 antibodies or nonspecific IgG, as indicated.
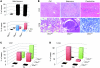
Figure 6. _Cbx7_-KO mice develop lung and liver tumors.
(A) Frequency of spontaneous solid tumor development in 17- to 22-month-old mice of each genotype (n = 11 [_Cbx7+/+_]; 34 [_Cbx7+/–_]; 24 [_Cbx7–/–_]). Ad, adenoma; Ca, carcinoma. (B) Representative liver and lung neoplasias in _Cbx7_-KO mice. Liver and lung tissues from Cbx7+/+ animals are shown on the left as normal controls. Original magnification, ×10. (C and D) Tumor incidence in livers (C) and lungs (D) from Cbx7+/+, Cbx7+/–, and Cbx7–/– mice.
Figure 7. Immunohistochemical analysis of CBX7 and cyclin E expression in human normal and neoplastic lung tissues.
(A and B) Normal lung had intense immunoreactivity for CBX7 (A, arrows), whereas it was negative for cyclin E expression (B). Arrowhead in B denotes positive histiocytes, a positive internal control. (C and D) Lung carcinoma was negative for CBX7 staining (C), but strongly positive for cyclin E expression (D). Higher-magnification views of A–D are provided in Supplemental Figure 7. (E and F) In a morphologically normal lung carcinoma–adjacent tissue, the tumoral part (top left) and the adjacent tissue (bottom right) did not express CBX7 (E), whereas both expressed high levels of cyclin E (F). Original magnification: ×63 (A–D); ×10 (E and F).
Similar articles
- Tumor suppressor activity of CBX7 in lung carcinogenesis.
Forzati F, Federico A, Pallante P, Fedele M, Fusco A. Forzati F, et al. Cell Cycle. 2012 May 15;11(10):1888-91. doi: 10.4161/cc.20022. Epub 2012 May 15. Cell Cycle. 2012. PMID: 22544325 - CBX7 is a glioma prognostic marker and induces G1/S arrest via the silencing of CCNE1.
Yu T, Wu Y, Hu Q, Zhang J, Nie E, Wu W, Wang X, Wang Y, Liu N. Yu T, et al. Oncotarget. 2017 Apr 18;8(16):26637-26647. doi: 10.18632/oncotarget.15789. Oncotarget. 2017. PMID: 28460453 Free PMC article. - Chromobox protein homologue 7 protein, with decreased expression in human carcinomas, positively regulates E-cadherin expression by interacting with the histone deacetylase 2 protein.
Federico A, Pallante P, Bianco M, Ferraro A, Esposito F, Monti M, Cozzolino M, Keller S, Fedele M, Leone V, Troncone G, Chiariotti L, Pucci P, Fusco A. Federico A, et al. Cancer Res. 2009 Sep 1;69(17):7079-87. doi: 10.1158/0008-5472.CAN-09-1542. Epub 2009 Aug 25. Cancer Res. 2009. PMID: 19706751 - CBX7 modulates the expression of genes critical for cancer progression.
Pallante P, Sepe R, Federico A, Forzati F, Bianco M, Fusco A. Pallante P, et al. PLoS One. 2014 May 27;9(5):e98295. doi: 10.1371/journal.pone.0098295. eCollection 2014. PLoS One. 2014. PMID: 24865347 Free PMC article. - CBX7 is Dualistic in Cancer Progression Based on its Function and Molecular Interactions.
Li J, Ouyang T, Li M, Hong T, Alriashy M, Meng W, Zhang N. Li J, et al. Front Genet. 2021 Oct 1;12:740794. doi: 10.3389/fgene.2021.740794. eCollection 2021. Front Genet. 2021. PMID: 34659360 Free PMC article. Review.
Cited by
- Genetic differences in transcript responses to low-dose ionizing radiation identify tissue functions associated with breast cancer susceptibility.
Snijders AM, Marchetti F, Bhatnagar S, Duru N, Han J, Hu Z, Mao JH, Gray JW, Wyrobek AJ. Snijders AM, et al. PLoS One. 2012;7(10):e45394. doi: 10.1371/journal.pone.0045394. Epub 2012 Oct 15. PLoS One. 2012. PMID: 23077491 Free PMC article. - CBX7 gene expression plays a negative role in adipocyte cell growth and differentiation.
Forzati F, Federico A, Pallante P, Colamaio M, Esposito F, Sepe R, Gargiulo S, Luciano A, Arra C, Palma G, Bon G, Bucher S, Falcioni R, Brunetti A, Battista S, Fedele M, Fusco A. Forzati F, et al. Biol Open. 2014 Sep 4;3(9):871-9. doi: 10.1242/bio.20147872. Biol Open. 2014. PMID: 25190058 Free PMC article. - Pluripotency and Epigenetic Factors in Mouse Embryonic Stem Cell Fate Regulation.
Morey L, Santanach A, Di Croce L. Morey L, et al. Mol Cell Biol. 2015 Aug;35(16):2716-28. doi: 10.1128/MCB.00266-15. Epub 2015 Jun 1. Mol Cell Biol. 2015. PMID: 26031336 Free PMC article. Review. - CBX7 regulates stem cell-like properties of gastric cancer cells via p16 and AKT-NF-κB-miR-21 pathways.
Ni SJ, Zhao LQ, Wang XF, Wu ZH, Hua RX, Wan CH, Zhang JY, Zhang XW, Huang MZ, Gan L, Sun HL, Dimri GP, Guo WJ. Ni SJ, et al. J Hematol Oncol. 2018 Feb 8;11(1):17. doi: 10.1186/s13045-018-0562-z. J Hematol Oncol. 2018. PMID: 29422082 Free PMC article. - miR-155 is positively regulated by CBX7 in mouse embryonic fibroblasts and colon carcinomas, and targets the KRAS oncogene.
Forzati F, De Martino M, Esposito F, Sepe R, Pellecchia S, Malapelle U, Pellino G, Arra C, Fusco A. Forzati F, et al. BMC Cancer. 2017 Mar 4;17(1):170. doi: 10.1186/s12885-017-3158-z. BMC Cancer. 2017. PMID: 28259135 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials