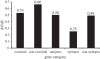Host-pathogen coevolution in human tuberculosis - PubMed (original) (raw)
Review
Host-pathogen coevolution in human tuberculosis
Sebastien Gagneux. Philos Trans R Soc Lond B Biol Sci. 2012.
Abstract
Tuberculosis (TB) is a disease of antiquity. Yet TB today still causes more adult deaths than any other single infectious disease. Recent studies show that contrary to the common view postulating an animal origin for TB, Mycobacterium tuberculosis complex (MTBC), the causative agent of TB, emerged as a human pathogen in Africa and colonized the world accompanying the Out-of-Africa migrations of modern humans. More recently, evolutionarily 'modern' lineages of MTBC expanded as a consequence of the global human population increase, and spread throughout the world following waves of exploration, trade and conquest. While epidemiological data suggest that the different phylogenetic lineages of MTBC might have adapted to different human populations, overall, the phylogenetically 'modern' MTBC lineages are more successful in terms of their geographical spread compared with the 'ancient' lineages. Interestingly, the global success of 'modern' MTBC correlates with a hypo-inflammatory phenotype in macrophages, possibly reflecting higher virulence, and a shorter latency in humans. Finally, various human genetic variants have been associated with different MTBC lineages, suggesting an interaction between human genetic diversity and MTBC variation. In summary, the biology and the epidemiology of human TB have been shaped by the long-standing association between MTBC and its human host.
Figures
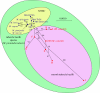
Figure 1.
Phylogenetic relationship between ‘smooth’ tubercle bacilli and classical MTBC. Multi-locus sequence data of six housekeeping genes show that the smooth tubercle bacilli are more genetically diverse than MTBC. The network structure of the phylogenetic relationships between members of the smooth tubercle bacilli indicates ongoing horizontal gene exchange, which is absent in MTBC. Adapted from Gutierrez et al. [19].
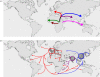
Figure 2.
Global phylogeography of MTBC. (a) Global phylogeny of human-adapted MTBC is based on 22 whole-genome sequences. Adapted from Comas et al. [35]. (b) Global distribution of the six main human-adapted MTBC lineages. Coloured dots represent the dominant MTBC lineage(s) in each country. Adapted from Gagneux et al. [36].
Figure 3.
‘Out-of-and-back-to-Africa’ scenario for the evolutionary history of human TB. Adapted from Hershberg et al. [32]. (a) MTBC originated in Africa and some lineages accompanied the Out-of-Africa migrations of modern humans. (b) The three evolutionary ‘modern’ MTBC lineages seeded Europe, India and China, respectively, and expanded as a consequence of the sharp increases in human populations in these regions starting a few centuries ago (each dark grey dot corresponds to 1 million people). These lineages then spread throughout the world via exploration, trade and conquest. Lineage colours correspond to figure 2.
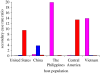
Figure 4.
Preferential sympatric transmission of MTBC lineages in San Francisco. The number of secondary TB cases generated between 2001 and 2009 is plotted as a function of the number of index cases (secondary case rate ratio) and the human population in which these secondary cases occurred. In each human population, the number of secondary cases caused by sympatric MTBC lineages was significantly higher compared with the secondary cases caused by allopatric lineages. Sympatric and allopatric lineages were defined based on their usual phylogeographic association shown in figure 2. (Red, lineage 4 or Euro-American lineage; Blue, lineage 2 or East-Asian lineage; Pink, lineage 1 or Indo-Oceanc lineage.) Adapted from Gagneux et al. [36].
Figure 5.
Human T-cell epitopes of MTBC are evolutionarily hyper-conserved. Ratio of non-synonymous mutations to synonymous mutations in different gene classes based on whole genome sequencing of 22 MTBC strains representative of the global MTBC diversity. Adapted from Comas et al. [35].
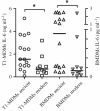
Figure 6.
Evolutionarily ‘modern’ MTBC lineages are hypo-inflammatory in human and murine macrophages. Human monocyte-derived (MDM) and murine bone marrow-derived (BMDM) macrophages were infected with one of 28 MTBC strains representative of the global MTBC diversity. Cytokines were measured in supernatants after 24 h of infection. *p < 0.05, Mann–Whitney U test. Adapted from Portevin et al. [57].
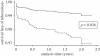
Figure 7.
Evolutionarily ‘modern’ MTBC lineages are associated with a shorter latency in human TB. TB patients of 1808 HIV-negative household contacts were followed over 2 years. The number of active TB cases occurring in these contacts during the study period was determined and stratified by the MTBC lineage of the index cases (solid line, ‘ancient’ MTBC (i.e. M. africanum), dashed line, ‘modern’ MTBC. Adapted from de Jong et al. [61].
Similar articles
- The Nature and Evolution of Genomic Diversity in the Mycobacterium tuberculosis Complex.
Brites D, Gagneux S. Brites D, et al. Adv Exp Med Biol. 2017;1019:1-26. doi: 10.1007/978-3-319-64371-7_1. Adv Exp Med Biol. 2017. PMID: 29116627 Review. - Clinical isolates of the modern Mycobacterium tuberculosis lineage 4 evade host defense in human macrophages through eluding IL-1β-induced autophagy.
Romagnoli A, Petruccioli E, Palucci I, Camassa S, Carata E, Petrone L, Mariano S, Sali M, Dini L, Girardi E, Delogu G, Goletti D, Fimia GM. Romagnoli A, et al. Cell Death Dis. 2018 May 24;9(6):624. doi: 10.1038/s41419-018-0640-8. Cell Death Dis. 2018. PMID: 29795378 Free PMC article. - Consequences of genomic diversity in Mycobacterium tuberculosis.
Coscolla M, Gagneux S. Coscolla M, et al. Semin Immunol. 2014 Dec;26(6):431-44. doi: 10.1016/j.smim.2014.09.012. Epub 2014 Oct 22. Semin Immunol. 2014. PMID: 25453224 Free PMC article. Review. - Shaping the niche in macrophages: Genetic diversity of the M. tuberculosis complex and its consequences for the infected host.
Reiling N, Homolka S, Kohl TA, Steinhäuser C, Kolbe K, Schütze S, Brandenburg J. Reiling N, et al. Int J Med Microbiol. 2018 Jan;308(1):118-128. doi: 10.1016/j.ijmm.2017.09.009. Epub 2017 Sep 14. Int J Med Microbiol. 2018. PMID: 28969988 Review. - Biological and Epidemiological Consequences of MTBC Diversity.
Coscolla M. Coscolla M. Adv Exp Med Biol. 2017;1019:95-116. doi: 10.1007/978-3-319-64371-7_5. Adv Exp Med Biol. 2017. PMID: 29116631 Review.
Cited by
- Whole genome sequencing of drug resistant Mycobacterium tuberculosis isolates from a high burden tuberculosis region of North West Pakistan.
Jabbar A, Phelan JE, de Sessions PF, Khan TA, Rahman H, Khan SN, Cantillon DM, Wildner LM, Ali S, Campino S, Waddell SJ, Clark TG. Jabbar A, et al. Sci Rep. 2019 Oct 18;9(1):14996. doi: 10.1038/s41598-019-51562-6. Sci Rep. 2019. PMID: 31628383 Free PMC article. - New developments in tuberculosis diagnosis and treatment.
Gill CM, Dolan L, Piggott LM, McLaughlin AM. Gill CM, et al. Breathe (Sheff). 2022 Mar;18(1):210149. doi: 10.1183/20734735.0149-2021. Epub 2021 Mar 8. Breathe (Sheff). 2022. PMID: 35284018 Free PMC article. Review. - Global MLST of Salmonella Typhi Revisited in Post-genomic Era: Genetic Conservation, Population Structure, and Comparative Genomics of Rare Sequence Types.
Yap KP, Ho WS, Gan HM, Chai LC, Thong KL. Yap KP, et al. Front Microbiol. 2016 Mar 2;7:270. doi: 10.3389/fmicb.2016.00270. eCollection 2016. Front Microbiol. 2016. PMID: 26973639 Free PMC article. - A single-nucleotide-polymorphism real-time PCR assay for genotyping of Mycobacterium tuberculosis complex in peri-urban Kampala.
Wampande EM, Hatzios SK, Achan B, Mupere E, Nsereko M, Mayanja HK, Eisenach K, Boom WH, Gagneux S, Joloba ML; Tuberculosis Research Unit. Wampande EM, et al. BMC Infect Dis. 2015 Sep 30;15:396. doi: 10.1186/s12879-015-1121-7. BMC Infect Dis. 2015. PMID: 26423522 Free PMC article. - Strains of Mycobacterium tuberculosis transmitting infection in Brazilian households and those associated with community transmission of tuberculosis.
Vinhas SA, Jones-López EC, Ribeiro Rodrigues R, Gaeddert M, Peres RL, Marques-Rodrigues P, de Aguiar PPL, White LF, Alland D, Salgame P, Hom D, Ellner JJ, Dietze R, Collins LF, Shashkina E, Kreiswirth B, Palaci M. Vinhas SA, et al. Tuberculosis (Edinb). 2017 May;104:79-86. doi: 10.1016/j.tube.2017.03.003. Epub 2017 Mar 16. Tuberculosis (Edinb). 2017. PMID: 28454653 Free PMC article.
References
- WHO 2010. Global tuberculosis control—surveillance, planning, financing. Geneva, Switzerland: WHO; (Contract No: WHO/HTM/TB/2008.393.)
- Barry C. E., 3rd, Boshoff H. I., Dartois V., Dick T., Ehrt S., Flynn J., Schnappinger D., Wilkinson R. J., Young D. 2009. The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nat. Rev. Microbiol. 7, 845–85510.1038/nrmicro2236 (doi:10.1038/nrmicro2236) - DOI - DOI - PMC - PubMed
- Wilson L. G. 1990. The historical decline of tuberculosis in Europe and America: its causes and significance. J. Hist. Med. Allied Sci. 45, 366–39610.1093/jhmas/45.3.366 (doi:10.1093/jhmas/45.3.366) - DOI - DOI - PubMed
- Dye C., Williams B. G. 2010. The population dynamics and control of tuberculosis. Science 328, 856–86110.1126/science.1185449 (doi:10.1126/science.1185449) - DOI - DOI - PubMed
- Lonnroth K., Jaramillo E., Williams B. G., Dye C., Raviglione M. 2009. Drivers of tuberculosis epidemics: the role of risk factors and social determinants. Soc. Sci. Med. 68, 2240–224610.1016/j.socscimed.2009.03.041 (doi:10.1016/j.socscimed.2009.03.041) - DOI - DOI - PubMed
Publication types
MeSH terms
Grants and funding
- MRC_U117588500/MRC_/Medical Research Council/United Kingdom
- AI090928/AI/NIAID NIH HHS/United States
- R01 AI090928/AI/NIAID NIH HHS/United States
- AI034238/AI/NIAID NIH HHS/United States
- R01 AI034238/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical