Safety assessment of Bifidobacterium longum JDM301 based on complete genome sequences - PubMed (original) (raw)
Safety assessment of Bifidobacterium longum JDM301 based on complete genome sequences
Yan-Xia Wei et al. World J Gastroenterol. 2012.
Abstract
Aim: To assess the safety of Bifidobacterium longum (B. longum) JDM301 based on complete genome sequences.
Methods: The complete genome sequences of JDM301 were determined using the GS 20 system. Putative virulence factors, putative antibiotic resistance genes and genes encoding enzymes responsible for harmful metabolites were identified by blast with virulence factors database, antibiotic resistance genes database and genes associated with harmful metabolites in previous reports. Minimum inhibitory concentration of 16 common antimicrobial agents was evaluated by E-test.
Results: JDM301 was shown to contain 36 genes associated with antibiotic resistance, 5 enzymes related to harmful metabolites and 162 nonspecific virulence factors mainly associated with transcriptional regulation, adhesion, sugar and amino acid transport. B. longum JDM301 was intrinsically resistant to ciprofloxacin, amikacin, gentamicin and streptomycin and susceptible to vancomycin, amoxicillin, cephalothin, chloramphenicol, erythromycin, ampicillin, cefotaxime, rifampicin, imipenem and trimethoprim-sulphamethoxazol. JDM301 was moderately resistant to bacitracin, while an earlier study showed that bifidobacteria were susceptible to this antibiotic. A tetracycline resistance gene with the risk of transfer was found in JDM301, which needs to be experimentally validated.
Conclusion: The safety assessment of JDM301 using information derived from complete bacterial genome will contribute to a wider and deeper insight into the safety of probiotic bacteria.
Keywords: Antibiotic resistance; Bifidobacterium longum; Genome; Harmful metabolite; Safety assessment; Virulence factor.
Figures
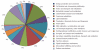
Figure 1
Functional distribution of Bifidobacterium longum core proteins. A total of 1265 proteins were conserved in all four Bifidobacterium longum (B. longum) strains (B. longum JDM301, B. longum NCC2705, B. longum DJO10A and B. longum ATCC15697), representing the “core” genome of B. longum.
Similar articles
- Safety Evaluations of Bifidobacterium bifidum BGN4 and Bifidobacterium longum BORI.
Kim MJ, Ku S, Kim SY, Lee HH, Jin H, Kang S, Li R, Johnston TV, Park MS, Ji GE. Kim MJ, et al. Int J Mol Sci. 2018 May 9;19(5):1422. doi: 10.3390/ijms19051422. Int J Mol Sci. 2018. PMID: 29747442 Free PMC article. - Antibiotic resistance of lactic acid bacteria and Bifidobacterium spp. isolated from dairy and pharmaceutical products.
D'Aimmo MR, Modesto M, Biavati B. D'Aimmo MR, et al. Int J Food Microbiol. 2007 Apr 1;115(1):35-42. doi: 10.1016/j.ijfoodmicro.2006.10.003. Epub 2007 Jan 2. Int J Food Microbiol. 2007. PMID: 17198739 - Complete genome sequence of Bifidobacterium longum JDM301.
Wei YX, Zhang ZY, Liu C, Zhu YZ, Zhu YQ, Zheng H, Zhao GP, Wang S, Guo XK. Wei YX, et al. J Bacteriol. 2010 Aug;192(15):4076-7. doi: 10.1128/JB.00538-10. Epub 2010 Jun 4. J Bacteriol. 2010. PMID: 20525832 Free PMC article. - Probiotics and safety.
Ishibashi N, Yamazaki S. Ishibashi N, et al. Am J Clin Nutr. 2001 Feb;73(2 Suppl):465S-470S. doi: 10.1093/ajcn/73.2.465s. Am J Clin Nutr. 2001. PMID: 11157359 Review. - Bifidobacterium longum W11: Uniqueness and individual or combined clinical use in association with rifaximin.
Di Pierro F, Pane M. Di Pierro F, et al. Clin Nutr ESPEN. 2021 Apr;42:15-21. doi: 10.1016/j.clnesp.2020.12.025. Epub 2021 Feb 17. Clin Nutr ESPEN. 2021. PMID: 33745570 Review.
Cited by
- Feasibility of Genome-Wide Screening for Biosafety Assessment of Probiotics: A Case Study of Lactobacillus helveticus MTCC 5463.
Senan S, Prajapati JB, Joshi CG. Senan S, et al. Probiotics Antimicrob Proteins. 2015 Dec;7(4):249-58. doi: 10.1007/s12602-015-9199-1. Probiotics Antimicrob Proteins. 2015. PMID: 26223907 - Addressing the Antibiotic Resistance Problem with Probiotics: Reducing the Risk of Its Double-Edged Sword Effect.
Imperial IC, Ibana JA. Imperial IC, et al. Front Microbiol. 2016 Dec 15;7:1983. doi: 10.3389/fmicb.2016.01983. eCollection 2016. Front Microbiol. 2016. PMID: 28018315 Free PMC article. Review. - Bifidobacterium adolescentis is intrinsically resistant to antitubercular drugs.
Lokesh D, Parkesh R, Kammara R. Lokesh D, et al. Sci Rep. 2018 Aug 9;8(1):11897. doi: 10.1038/s41598-018-30429-2. Sci Rep. 2018. PMID: 30093677 Free PMC article. - Safety Evaluation and Colonisation Abilities of Four Lactic Acid Bacteria as Future Probiotics.
Dlamini ZC, Langa RLS, Aiyegoro OA, Okoh AI. Dlamini ZC, et al. Probiotics Antimicrob Proteins. 2019 Jun;11(2):397-402. doi: 10.1007/s12602-018-9430-y. Probiotics Antimicrob Proteins. 2019. PMID: 29881953 - Safety Assessment of Lactobacillus helveticus KLDS1.8701 Based on Whole Genome Sequencing and Oral Toxicity Studies.
Li B, Jin D, Etareri Evivie S, Li N, Yan F, Zhao L, Liu F, Huo G. Li B, et al. Toxins (Basel). 2017 Sep 24;9(10):301. doi: 10.3390/toxins9100301. Toxins (Basel). 2017. PMID: 28946645 Free PMC article.
References
- Marco ML, Pavan S, Kleerebezem M. Towards understanding molecular modes of probiotic action. Curr Opin Biotechnol. 2006;17:204–210. - PubMed
- Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, Whitehead TR, Lapidus A, Rokhsar DS, Lebrilla CB, German JB, et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci USA. 2008;105:18964–18969. - PMC - PubMed
- Borriello SP, Hammes WP, Holzapfel W, Marteau P, Schrezenmeir J, Vaara M, Valtonen V. Safety of probiotics that contain lactobacilli or bifidobacteria. Clin Infect Dis. 2003;36:775–780. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials