Isolation and genomic analysis of circulating tumor cells from castration resistant metastatic prostate cancer - PubMed (original) (raw)
Isolation and genomic analysis of circulating tumor cells from castration resistant metastatic prostate cancer
Mark Jesus M Magbanua et al. BMC Cancer. 2012.
Abstract
Background: The number of circulating tumor cells (CTCs) in metastatic prostate cancer patients provides prognostic and predictive information. However, it is the molecular characterization of CTCs that offers insight into the biology of these tumor cells in the context of personalized treatment.
Methods: We developed a novel approach to isolate CTCs away from hematopoietic cells with high purity, enabling genomic analysis of these cells. The isolation protocol involves immunomagnetic enrichment followed by fluorescence activated cell sorting (IE/FACS). To evaluate the feasibility of isolation of CTCs by IE/FACS and downstream genomic profiling, we conducted a pilot study in patients with metastatic castration resistant prostate cancer (CRPC). Twenty (20) sequential CRPC patients were assayed using CellSearch™. Twelve (12) patients positive for CTCs were subjected to immunomagnetic enrichment and fluorescence activated cell sorting (IE/FACS) to isolate CTCs. Genomic DNA of CTCs was subjected to whole genome amplification (WGA) followed by gene copy number analysis via array comparative genomic hybridization (aCGH).
Results: CTCs from nine (9) patients successfully profiled were observed to have multiple copy number aberrations including those previously reported in primary prostate tumors such as gains in 8q and losses in 8p. High-level copy number gains at the androgen receptor (AR) locus were observed in 7 (78%) cases. Comparison of genomic profiles between CTCs and archival primary tumors from the same patients revealed common lineage. However, high-level copy number gains in the AR locus were observed in CTCs, but not in the matched archival primary tumors.
Conclusions: We developed a new approach to isolate prostate CTCs without significant leukocyte admixture, and to subject them to genome-wide copy number analysis. Our assay may be utilized to explore genomic events involved in cancer progression, e.g. development of castration resistance and to monitor therapeutic efficacy of targeted therapies in clinical trials in a relatively non-invasive manner.
Figures
Figure 1
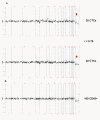
Copy number analysis of spiked cells isolated from healthy blood via IE/FACS. Twenty (20) isolated (A) LNCaP, (B) PC3 and (C) VCaP cells (in duplicate) and 50 ng of genomic DNA from cell culture subjected to whole genome amplification, and unamplified genomic DNA (600 ng) from cell culture (positive control) were analyzed for copy number aberrations. The log2 ratio value for each BAC clone is plotted on the y-axis. The x-axis represents the genomic position of each BAC clone on the array, with chromosome numbers indicated. Vertical solid lines indicate chromosome boundaries, and vertical red dashed line represents the centromeric region dividing each chromosome into the p- or short arm (to the left of centromere) and the q- or long arm (to the right of the centromere). Arrows indicate high-level gains on X chromosome region containing AR observed in VCaP cells.
Figure 2
Copy number analysis of CTCs. Array comparative genomic hybridization analysis of (A) 20 CTCs in duplicate (B) and 100 leukocytes (CD45-positive) isolated from the same enriched blood sample from patient #9. The log2 ratio value for each BAC clone is plotted on the y-axis. The x-axis represents the genomic position of each BAC clone on the array, with chromosome numbers indicated. Vertical solid lines indicate chromosome boundaries, and vertical red dashed line represents the centromeric region dividing each chromosome into the p- or short arm (to the left of centromere) and the q- or long arm (to the right of the centromere). Arrows indicate high-level gains on X chromosome region containing AR.
Figure 3
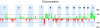
Frequency of copy number alterations in CTCs from 9 metastatic CRPC patients. Gains and losses are shown in green and red, respectively. Chromosome Y was not included in the analysis.
Figure 4
X chromosome and AR locus copy number analysis in CTCs from 9 metastatic CRPC patients. Each plot shows log2 hybridization ratio values (y-axis) at BAC clones (blue dots) distributed from the p terminus to the q terminus (x-axis). The black horizontal lines represent log2 ratio equal to 0. Red vertical lines demarcate the centromere. Broken black lines superimposed on BAC clones are the output of segmentation analysis providing high confidence copy number calls. Transparent blue bars identify the locus containing the androgen receptor gene (AR). At the bottom of each panel is an ideogram of chromosome X with cytoband regions showing gains (green) and losses (red). Bars on top of the ideogram (long arm) show results of copy number analysis at the AR locus (double green bar- high-level copy number gain; single green bar- low level copy number gain, grey bar- no copy number change). Paired samples are enclosed in black boxes: CTCs vs. CD45-positive (leukocytes) from patient #9 and CTCs vs. archival primary tumor (PT) from patients #17 and #20.
Figure 5
Copy number analysis of CTCs versus matched archival tumor in two patients. Array CGH analysis of (A) 20 CTCs from patient #17 and a corresponding archival primary tumor from local extension to the bladder obtained 1 year and 3 months prior to CTC analysis; and (B) 18 CTCs from patient #20 and a matched primary tumor obtained 5 years and 1 month prior to CTC analysis. Arrows indicate high-level gains on X chromosome region containing AR in CTCs but not observed in archival primary tumors. Chromosome regions in red boxes show genomic aberrations common to both paired samples (Also see Additional file 3: Figure S3).
Similar articles
- Genomic profiling of isolated circulating tumor cells from metastatic breast cancer patients.
Magbanua MJ, Sosa EV, Roy R, Eisenbud LE, Scott JH, Olshen A, Pinkel D, Rugo HS, Park JW. Magbanua MJ, et al. Cancer Res. 2013 Jan 1;73(1):30-40. doi: 10.1158/0008-5472.CAN-11-3017. Epub 2012 Nov 7. Cancer Res. 2013. PMID: 23135909 Free PMC article. - Circulating tumor cell analysis in patients with progressive castration-resistant prostate cancer.
Shaffer DR, Leversha MA, Danila DC, Lin O, Gonzalez-Espinoza R, Gu B, Anand A, Smith K, Maslak P, Doyle GV, Terstappen LW, Lilja H, Heller G, Fleisher M, Scher HI. Shaffer DR, et al. Clin Cancer Res. 2007 Apr 1;13(7):2023-9. doi: 10.1158/1078-0432.CCR-06-2701. Clin Cancer Res. 2007. PMID: 17404082 - Whole Genomic Copy Number Alterations in Circulating Tumor Cells from Men with Abiraterone or Enzalutamide-Resistant Metastatic Castration-Resistant Prostate Cancer.
Gupta S, Li J, Kemeny G, Bitting RL, Beaver J, Somarelli JA, Ware KE, Gregory S, Armstrong AJ. Gupta S, et al. Clin Cancer Res. 2017 Mar 1;23(5):1346-1357. doi: 10.1158/1078-0432.CCR-16-1211. Epub 2016 Sep 6. Clin Cancer Res. 2017. PMID: 27601596 - Circulating tumor cells as biomarkers in prostate cancer.
Danila DC, Fleisher M, Scher HI. Danila DC, et al. Clin Cancer Res. 2011 Jun 15;17(12):3903-12. doi: 10.1158/1078-0432.CCR-10-2650. Clin Cancer Res. 2011. PMID: 21680546 Free PMC article. Review. - Clinical significance of genomic sequencing of circulating tumour cells (CTCs) in cancer.
Auwal A, Hossain MM, Pronoy TUH, Rashel KM, Nurujjaman M, Lam AK, Islam F. Auwal A, et al. J Liq Biopsy. 2023 Dec 28;3:100135. doi: 10.1016/j.jlb.2023.100135. eCollection 2024 Mar. J Liq Biopsy. 2023. PMID: 40026568 Free PMC article. Review.
Cited by
- Experimental evidence of persistent androgen-receptor-dependency in castration-resistant prostate cancer.
Kobayashi T, Inoue T, Kamba T, Ogawa O. Kobayashi T, et al. Int J Mol Sci. 2013 Jul 26;14(8):15615-35. doi: 10.3390/ijms140815615. Int J Mol Sci. 2013. PMID: 23896594 Free PMC article. Review. - A comparison of isolated circulating tumor cells and tissue biopsies using whole-genome sequencing in prostate cancer.
Jiang R, Lu YT, Ho H, Li B, Chen JF, Lin M, Li F, Wu K, Wu H, Lichterman J, Wan H, Lu CL, OuYang W, Ni M, Wang L, Li G, Lee T, Zhang X, Yang J, Rettig M, Chung LW, Yang H, Li KC, Hou Y, Tseng HR, Hou S, Xu X, Wang J, Posadas EM. Jiang R, et al. Oncotarget. 2015 Dec 29;6(42):44781-93. doi: 10.18632/oncotarget.6330. Oncotarget. 2015. PMID: 26575023 Free PMC article. - KRAS genotypic changes of circulating tumor cells during treatment of patients with metastatic colorectal cancer.
Kalikaki A, Politaki H, Souglakos J, Apostolaki S, Papadimitraki E, Georgoulia N, Tzardi M, Mavroudis D, Georgoulias V, Voutsina A. Kalikaki A, et al. PLoS One. 2014 Aug 19;9(8):e104902. doi: 10.1371/journal.pone.0104902. eCollection 2014. PLoS One. 2014. PMID: 25137394 Free PMC article. - Circulating tumour cells-monitoring treatment response in prostate cancer.
Miyamoto DT, Sequist LV, Lee RJ. Miyamoto DT, et al. Nat Rev Clin Oncol. 2014 Jul;11(7):401-12. doi: 10.1038/nrclinonc.2014.82. Epub 2014 May 13. Nat Rev Clin Oncol. 2014. PMID: 24821215 Review. - Recent advances in the biology of human circulating tumour cells and metastasis.
Gkountela S, Szczerba B, Donato C, Aceto N. Gkountela S, et al. ESMO Open. 2016 Aug 3;1(4):e000078. doi: 10.1136/esmoopen-2016-000078. eCollection 2016. ESMO Open. 2016. PMID: 27843628 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials