The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome - PubMed (original) (raw)
The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome
Alex J Kuo et al. Nature. 2012.
Abstract
The recognition of distinctly modified histones by specialized 'effector' proteins constitutes a key mechanism for transducing molecular events at chromatin to biological outcomes. Effector proteins influence DNA-templated processes, including transcription, DNA recombination and DNA repair; however, no effector functions have yet been identified within the mammalian machinery that regulate DNA replication. Here we show that ORC1--a component of ORC (origin of replication complex), which mediates pre-DNA replication licensing--contains a bromo adjacent homology (BAH) domain that specifically recognizes histone H4 dimethylated at lysine 20 (H4K20me2). Recognition of H4K20me2 is a property common to BAH domains present within diverse metazoan ORC1 proteins. Structural studies reveal that the specificity of the BAH domain for H4K20me2 is mediated by a dynamic aromatic dimethyl-lysine-binding cage and multiple intermolecular contacts involving the bound peptide. H4K20me2 is enriched at replication origins, and abrogating ORC1 recognition of H4K20me2 in cells impairs ORC1 occupancy at replication origins, ORC chromatin loading and cell-cycle progression. Mutation of the ORC1 BAH domain has been implicated in the aetiology of Meier-Gorlin syndrome (MGS), a form of primordial dwarfism, and ORC1 depletion in zebrafish results in an MGS-like phenotype. We find that wild-type human ORC1, but not ORC1-H4K20me2-binding mutants, rescues the growth retardation of orc1 morphants. Moreover, zebrafish depleted of H4K20me2 have diminished body size, mirroring the phenotype of orc1 morphants. Together, our results identify the BAH domain as a novel methyl-lysine-binding module, thereby establishing the first direct link between histone methylation and the metazoan DNA replication machinery, and defining a pivotal aetiological role for the canonical H4K20me2 mark, via ORC1, in primordial dwarfism.
Figures
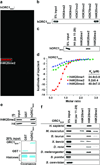
Figure 1. The ORC1 BAH domain is a novel H4K20me2-binding module
a, hORC1BAH preferentially binds H4K20me2 peptides. Microarrays spotted with 82 distinct histone peptides as indicated in Supplementary Fig. 1 were probed with glutathione S-transferase (GST) fused to human ORC1 amino acids (aa) 1-185 (hORC1BAH). Red spots indicate positive binding. b and c, hORC1BAH specifically binds H4K20me2 peptides. Western blot analysis of histone peptide pull-downs with GST-hORC1BAH and the indicated biotinylated peptides. d, hORC1BAH binds with highest affinity to H4K20me2. ITC was used to determine the dissociation constants (_K_ds) for the interaction of hORC1BAH with the indicated peptides: H4K20me1 (green diamonds), H4K20me2 (red squares) and H4K20me3 (blue triangles). Standard deviation (s.d.) was derived from nonlinear fitting. e, hORC1BAH binds full-length H4K20me2. Top: Western blot of GST-hORC1BAH and GST control pull-downs from calf thymus histones (CTH) with the indicated antibodies. Bottom: Coomassie blue stain of input (20% of total). f, H4K20me2 binding is a common property of ORC1 BAH domains from diverse metazoan species. Binding assays as in (b) with GST-fused ORC1 BAH domains from the indicated species and using the indicated peptides.
Figure 2. The molecular basis of H4K20me2 recognition by ORC1BAH
a–c, 1.95 Å crystal structure of murine ORC1 BAH aa 9-170 (mORC1BAH) complexed with H4(14-25)K20me2 peptide. a, Ribbon representation of mORC1BAH bound to H4K20me2 peptide. The mORC1BAH (cyan) and the bound H4K20me2 peptide (yellow) are shown in ribbon and stick representations, respectively. b, Details of intermolecular contacts in the mORC1BAH-H4K20me2 complex. mORC1BAH and H4K20me2 peptide residues are colored in pink and yellow, respectively, with hydrogen bonds depicted as red dashed lines, and a water molecular as a red sphere. c, Schematic representation of intermolecular contacts in the mORC1BAH-H4K20me2 complex. The residues from the H4K20me2 peptide and mORC1BAH are colored in magenta and black, respectively. Yellow: the hydrophobic contact. d, Positioning of the K20me2 side chain within an aromatic cage of the indicated residues (red) on the surface of mORC1BAH. The equivalent cage residues in hORC1BAH are labeled in parenthesis. e, Structural overlay of the mORC1BAH K20me2-binding aromatic cage in the free (silver) and H4K20me2-bound states (salmon). Arrows indicate binding-induced structural shifts. f, Mutations in the hORC1BAH H4K20me2-binding channel impair H4K20me2 recognition. ITC analysis of the indicated hORC1BAH mutants binding to H4K20me2 peptide; s.d. derived from non-linear fitting. g–h, Mutations in the hORC1BAH dimethyllysine-binding cage abrogate H4K20me2 recognition. g, Binding assays as in (Fig. 1b) with the indicated hORC1BAH mutant proteins and biotinylated peptides. h, Top: Western analysis of CTH binding assays as in (Fig. 1e) with the indicated proteins and antibodies. Bottom: Coomassie blue stain of input GST-fusion proteins and histones (20% of total).
Figure 3. ORC1-H4K20me2 interaction regulates ORC chromatin association and cell cycle progression
a, Western blot analysis with the indicated antibodies of wild-type (WT) and H4K20me2-binding pocket mutants (Y64A and W88A) affinity-purified FLAG-tagged ORC1 complexes from U2OS cells. Control, empty vector control IP. WCE, whole cell extract. b, The ORC1BAH-H4K20me2 interaction is required for efficient ORC1 chromatin association. Western blot analysis of lysates biochemically separated into chromatin-enriched and soluble fractions from U2OS cells stably expressing the indicated ORC1 protein. Quantitation of FLAG-ORC1 levels is shown. Control, empty vector control lysates. Tubulin and H3 levels are shown as control for the integrity of fractionation. c, Disruption of ORC1 binding to H4K20me2 destabilizes ORC chromatin association. Western blot analysis of biochemically purified chromatin from U2OS cells as in (b) with the indicated antibodies. Total ORC protein levels in WCE are shown. d, H4K20me2 is enriched at DNA replication origins. H4K20me2 signal normalized to total H4 at the MCM4 origin and indicated flanking regions in G1 phase synchronized U2OS cells stably expressing the indicated ORC protein; _y_-axis: H4K20me2 ChIP/H4 ChIP. e, An intact BAH domain is required for ORC1 occupancy at replication origins. Occupancy of FLAG tagged hORC1, hORC1Y64A, hORC1W88A, or control was determined by ChIP analysis (_y_-axs: % input) as in (d). Error bars in (d, and e,) indicate s.e.m. from three experiments. f, The ORC1-H4K20me2 interaction is required for efficient cell cycle progression. The cell-cycle profile of WI38 cells transiently expressing GFP and the indicated ORC1 protein was determined by flow cytometry. GFP-positive cells were used to ensure that only transfected cells were analyzed. (Left) Percentage of cells in the indicated cell-cycle phase is shown. (Right) The S/G1 ratio for the indicated transfections is shown. Error bars indicate s.d. from two experiments.
Figure 4. Disruption of the ORC1BAH-H4K20me2 interaction leads to dwarfism in zebrafish
a–c, MGS-associated mutations F89S and E127G impair H4K20me2 binding by hORC1. a, Close-up view of mORC1BAH bound to H4K20me2, with residues F88 and E126 (equivalent to F89 and E127 in hORC1, respectively) shown in stick representation. b, ITC analysis as in (Fig. 1d) of F89S and E127G hORC1 BAH mutants binding to H4K20me2 peptides. *ITC data for wild-type hORC1BAH from Fig. 1d. c, Binding assays as in (Fig. 1b) with the indicated hORC1BAH mutant proteins and biotinylated peptides. d–e, The H4K20me2-recognition activity of hORC1 is required to rescue the dwarfism phenotype of orc1 morphants. d, Quantification of dwarf phenotype in zebrafish injected with morphilino oligos (MO) targeting the orc1 translation start site (tss) alone or MO co-injected with the indicated hORC1 mRNAs. Insert on left shows electrophoresis analysis of the indicated hORC1 mRNAs used for reconstitution. Control, uninjected embryos. Dwarfism was defined as a reduction of body length of ≥ 3 s.d. relative to the average size of the control zebrafish. Zebrafish analyzed: control: 26; orc1 MO: 86; MO+hORC1WT: 67; MO+hORC1Y64A: 85; MO+hORC1W88A: 83. P values are calculated with a two-tailed unpaired Student’s _t_-test. ** P < 0.01. e, Representative images of zebrafish in (b) 1 day post-fertilization (dpf). f, Depletion of H4K20me2/3 in suv4-20h1/suv4-20h2 morphants. Western blot analysis using the indicated antibodies of whole animal extracts 1 dpf from control zebrafish or zebrafish injected with MOs targeting the tss’s of suv4-20h1 and suv4-20h2. Control, uninjected embryos. g–h, Dwarfism in suv4-20h1/suv4-20h2 morphants. g, Quantification of dwarfism in suv4-20h1/suv4-20h2 morphants relative to controls as described in (d). Zebrafish analyzed: control: 38; MO: 60. ** P<0.01. h, representative images of zebrafish in (e) 1 dpf.
Similar articles
- Mutations in ORC1, encoding the largest subunit of the origin recognition complex, cause microcephalic primordial dwarfism resembling Meier-Gorlin syndrome.
Bicknell LS, Walker S, Klingseisen A, Stiff T, Leitch A, Kerzendorfer C, Martin CA, Yeyati P, Al Sanna N, Bober M, Johnson D, Wise C, Jackson AP, O'Driscoll M, Jeggo PA. Bicknell LS, et al. Nat Genet. 2011 Feb 27;43(4):350-5. doi: 10.1038/ng.776. Nat Genet. 2011. PMID: 21358633 - A Meier-Gorlin syndrome mutation impairs the ORC1-nucleosome association.
Zhang W, Sankaran S, Gozani O, Song J. Zhang W, et al. ACS Chem Biol. 2015 May 15;10(5):1176-80. doi: 10.1021/cb5009684. Epub 2015 Feb 24. ACS Chem Biol. 2015. PMID: 25689043 Free PMC article. - Deficiency in origin licensing proteins impairs cilia formation: implications for the aetiology of Meier-Gorlin syndrome.
Stiff T, Alagoz M, Alcantara D, Outwin E, Brunner HG, Bongers EM, O'Driscoll M, Jeggo PA. Stiff T, et al. PLoS Genet. 2013;9(3):e1003360. doi: 10.1371/journal.pgen.1003360. Epub 2013 Mar 14. PLoS Genet. 2013. PMID: 23516378 Free PMC article. - The origin recognition complex in human diseases.
Shen Z. Shen Z. Biosci Rep. 2013 Jun 11;33(3):e00044. doi: 10.1042/BSR20130036. Biosci Rep. 2013. PMID: 23662735 Free PMC article. Review. - Meier-Gorlin syndrome.
de Munnik SA, Hoefsloot EH, Roukema J, Schoots J, Knoers NV, Brunner HG, Jackson AP, Bongers EM. de Munnik SA, et al. Orphanet J Rare Dis. 2015 Sep 17;10:114. doi: 10.1186/s13023-015-0322-x. Orphanet J Rare Dis. 2015. PMID: 26381604 Free PMC article. Review.
Cited by
- Crossing epigenetic frontiers: the intersection of novel histone modifications and diseases.
Yao W, Hu X, Wang X. Yao W, et al. Signal Transduct Target Ther. 2024 Sep 16;9(1):232. doi: 10.1038/s41392-024-01918-w. Signal Transduct Target Ther. 2024. PMID: 39278916 Free PMC article. Review. - Replication licensing regulated by a short linear motif within an intrinsically disordered region of origin recognition complex.
Wu Y, Zhang Q, Lin Y, Lam WH, Zhai Y. Wu Y, et al. Nat Commun. 2024 Sep 13;15(1):8039. doi: 10.1038/s41467-024-52408-0. Nat Commun. 2024. PMID: 39271725 Free PMC article. - Histone variant macroH2A1 regulates synchronous firing of replication origins in the inactive X chromosome.
Arroyo M, Casas-Delucchi CS, Pabba MK, Prorok P, Pradhan SK, Rausch C, Lehmkuhl A, Maiser A, Buschbeck M, Pasque V, Bernstein E, Luck K, Cardoso MC. Arroyo M, et al. Nucleic Acids Res. 2024 Oct 28;52(19):11659-11688. doi: 10.1093/nar/gkae734. Nucleic Acids Res. 2024. PMID: 39189450 Free PMC article. - Single-electron transfer between sulfonium and tryptophan enables site-selective photo crosslinking of methyllysine reader proteins.
Feng F, Gao Y, Zhao Q, Luo T, Yang Q, Zhao N, Xiao Y, Han Y, Pan J, Feng S, Zhang L, Wu M. Feng F, et al. Nat Chem. 2024 Aug;16(8):1267-1277. doi: 10.1038/s41557-024-01577-y. Epub 2024 Jul 30. Nat Chem. 2024. PMID: 39079947 - Determinants of Chromatin Organization in Aging and Cancer-Emerging Opportunities for Epigenetic Therapies and AI Technology.
Castilho RM, Castilho LS, Palomares BH, Squarize CH. Castilho RM, et al. Genes (Basel). 2024 May 29;15(6):710. doi: 10.3390/genes15060710. Genes (Basel). 2024. PMID: 38927646 Free PMC article. Review.
References
- Bicknell LS, et al. Mutations in ORC1, encoding the largest subunit of the origin recognition complex, cause microcephalic primordial dwarfism resembling Meier-Gorlin syndrome. Nat Genet. 2011;43:350–355. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- DP1 HD075622/HD/NICHD NIH HHS/United States
- DP1 OD003792-04/OD/NIH HHS/United States
- DP1 OD003792/OD/NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- R01 GM079641/GM/NIGMS NIH HHS/United States
- R01GM079641/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases