ESCRT-III governs the Aurora B-mediated abscission checkpoint through CHMP4C - PubMed (original) (raw)
ESCRT-III governs the Aurora B-mediated abscission checkpoint through CHMP4C
Jeremy G Carlton et al. Science. 2012.
Abstract
The endosomal sorting complex required for transport (ESCRT) machinery plays an evolutionarily conserved role in cytokinetic abscission, the final step of cell division where daughter cells are physically separated. Here, we show that charged multivesicular body (MVB) protein 4C (CHMP4C), a human ESCRT-III subunit, is involved in abscission timing. This function correlated with its differential spatiotemporal distribution during late stages of cytokinesis. Accordingly, CHMP4C functioned in the Aurora B-dependent abscission checkpoint to prevent both premature resolution of intercellular chromosome bridges and accumulation of DNA damage. CHMP4C engaged the chromosomal passenger complex (CPC) via interaction with Borealin, which suggested a model whereby CHMP4C inhibits abscission upon phosphorylation by Aurora B. Thus, the ESCRT machinery may protect against genetic damage by coordinating midbody resolution with the abscission checkpoint.
Figures
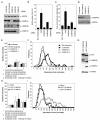
Figure 1. CHMP4C negatively regulates cytokinesis
A. Resolved HeLa cell lysates were examined by blotting with a α-CHMP4A, α-CHMP4B, α-CHMP4C or α-Heat Shock Protein 90 kDa (HSP90). B, C. siRNA-transfected HeLa cells were fixed and stained with α-Tubulin. Multinucleate cells (B, n=3±SD) or cells connected by midbodies (C, n=7±SD) were scored visually. D. Resolved lysates from siRNA-transfected HeLa mCh-Tub cells were examined by western blotting with α-CHMP4C or α-HSP90. E-F. Asynchronous cultures of HeLa mCh-Tub cells were transfected with the indicated siRNA and imaged live and mitotic durations quantified. Abscission time (Luciferase: 93±38 minutes, n = 96; Non-targeting: 94±36 minutes, n = 94; CHMP4C-1: 59±17 minutes, n = 88; CHMP4C-2: 61±25 minutes, n = 100) was calculated across 4 independent experiments. G. Resolved cell lysates from HeLa cells stably expressing mCh-Tubulin and either GFP-CHMP4B or GFP-CHMP4C were examined by blotting with α-HSP90, α-GFP, α-CHMP4B or α-CHMP4C. H, I. Cells from G were imaged live and mitotic durations quantified. The more intense imaging (17) resulted in general abscission delays to 116±45 and 137±61 minutes for control or GFP-CHMP4B expressing cells whilst GFP-CHMP4C expressing cells took 240±103 minutes to complete abscission. Data comprises 185 cells per condition from 3 independent experiments.
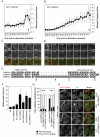
Figure 2. Differential spatio-temporal recruitment of CHMP4 paralogs during cytokinesis
A-D. GFP-fluorescence intensities of midbody-localised GFP-CHMP4B (A, C n=14±S.D.) or GFP-CHMP4C (B, D n=9±S.D.) during abscission. Abscission indicated by arrow, time in minutes, selected frames presented (C, D). Initial recruitment of GFP-CHMP4C to midbody arms marked (arrowhead). E. ClustalW alignment of the C-terminal regions of CHMP4A, CHMP4B and CHMP4C, S210 indicated by arrow. F. HeLa cells transfected with plasmids encoding the indicated HA-CHMP4 constructs were fixed and stained with α-Tubulin and α-HA. Multinucleate cells were scored (n=5±S.D.). G, H. HeLa mCh-Tub cells stably expressing HA-CHMP4CR, HA-CHMP4CRδINS or HA-CHMP4CR S210A were treated with CHMP4C siRNA, fixed, stained with α-HA and HA-CHMP4C location was scored (n=3±S.D.) Bar is 10 μm.
Figure 3. CHMP4C regulates the abscission checkpoint
A-C. Cell lysates from siRNA-transfected HeLa cells were examined by blotting with α-NUP153, α-CHMP4C, α-HA and α-HSP90 (A). Alternatively, cells were fixed and stained with α-tubulin (B) or α-tubulin and α-pT232 Aurora B (C) Bar is 10 μm. Multinucleate and midbody-connected cells were scored visually (A, n=6±S.D.). D, E. HeLa cells stably expressing YFP-LAP2β, were transfected with plasmids encoding the indicated HA-CHMP4 constructs. Cells were fixed, stained with α-Tubulin and α-HA (D), or α-pT232 Aurora B and α-HA (E) Bar is 10 μm. F. HeLa cells stably expressing YFP-LAP2β were transfected with the indicated siRNA, imaged live and the duration of LAP2β-bridge resolution (Luciferase: 576±454 minutes, n = 116; CHMP4C-1: 321±308 minutes, n = 112; CHMP4C-2: 291±278 minutes, n = 103) was quantified across 6 independent experiments G. Cell lysates from clonal shRNA-transduced HeLa cells were examined by blotting with α-γH2AX, α-CHMP4C or α-HSP90.
Figure 4. Aurora B-dependent phosphorylation of CHMP4C S210 activates the NoCut abscission checkpoint
A. β-galactosidase assay from yeast co-transformed with the indicated VP16 and GAL4-fused constructs (n=3±S.D.). B. Cell lysates and glutathione-bound fractions from 293T cells transfected with the indicated fusion proteins were examined by western blotting with α-HA. C. HeLa mCh-Tub cells stably expressing HA-CHMP4CR were fixed and stained with α-HA and α-Aurora B. D. Asynchronous and mitotic lysates of HeLa mCh-Tub cells stably expressing HA-CHMP4CR were immunoprecipitated with α–HA and treated as indicated and examined by blotting with α–HA, α-CHMP4C, α-CHMP4B, α–CEP55 and α–HSP90. E. F. Asynchronous or mitotically arrested HeLa mCh-Tub cells stably expressing HA-CHMP4CR were either released into media containing DMSO or the phosphatase inhbitor Okadaic Acid (OA) for the indicated times (E) or were treated overnight during the nocodazole arrest with inhibitors of MEK (U0126), PI 3-kinase (LY294002) or Aurora B (ZM447439) (F). Cell lysates were examined by blotting with α–HA and α–HSP90. G. H. Proteins were immunoprecipitated from 293T cells with α–HA and subjected to an in-vitro kinase assay with recombinant Aurora B. Incorporated 32P was visualized by phosphorimaging, blotting with α–HA allowed detection of immunoprecipitates. I-K. Asynchronous cultures of HeLa mCh-Tub cells stably expressing HA, HA-CHMP4CR, HA-CHMP4CR δINS, or HA-CHMP4CR S210A were transfected with the indicated siRNA. Resolved cell lysates were examined by blotting with α-CHMP4C, α-HA and α-HSP90 (I). Alternatively, cells were imaged live (J, K) and abscission time (Luciferase, 104±35 minutes, n = 244, CHMP4c siRNA, 71±37 minutes, n = 260; CHMP4c siRNA and HA-CHMP4CR, 118±52 minutes, n = 269; CHMP4C siRNA and HA-CHMP4CR δINS, 81±40 minutes, n = 264; CHMP4C siRNA and HA-CHMP4CR S210A, 91±38 minutes, n = 268) quantified across 7 independent experiments. L, HeLa cells stably expressing YFP-LAP2β and either HA or HA-CHMP4CR, HA-CHMP4CR δINS, or HA-CHMP4CR S210A were treated with the indicated siRNA, imaged live and the timing of YFP-LAP2β bridge resolution (Luciferase, 628 ± 382 minutes, n = 41; CHMP4c siRNA, 413 ± 292 minutes, n = 41; CHMP4c siRNA and HA-CHMP4CR, 698 ± 332 minutes, n = 41; CHMP4C siRNA and HA-CHMP4CR δINS, 402 ± 259 minutes, n = 36; CHMP4C siRNA and HA-CHMP4CR S210A, 421 ± 295 minutes, n = 40) quantified from 2 independent experiments.
Comment in
- Cell biology. ESCRTing DNA at the cleavage site during cytokinesis.
Petronczki M, Uhlmann F. Petronczki M, et al. Science. 2012 Apr 13;336(6078):166-7. doi: 10.1126/science.1221832. Science. 2012. PMID: 22499931 No abstract available.
Similar articles
- Coordinated regulation of the ESCRT-III component CHMP4C by the chromosomal passenger complex and centralspindlin during cytokinesis.
Capalbo L, Mela I, Abad MA, Jeyaprakash AA, Edwardson JM, D'Avino PP. Capalbo L, et al. Open Biol. 2016 Oct;6(10):160248. doi: 10.1098/rsob.160248. Open Biol. 2016. PMID: 27784789 Free PMC article. - The chromosomal passenger complex controls the function of endosomal sorting complex required for transport-III Snf7 proteins during cytokinesis.
Capalbo L, Montembault E, Takeda T, Bassi ZI, Glover DM, D'Avino PP. Capalbo L, et al. Open Biol. 2012 May;2(5):120070. doi: 10.1098/rsob.120070. Open Biol. 2012. PMID: 22724069 Free PMC article. - ULK3 regulates cytokinetic abscission by phosphorylating ESCRT-III proteins.
Caballe A, Wenzel DM, Agromayor M, Alam SL, Skalicky JJ, Kloc M, Carlton JG, Labrador L, Sundquist WI, Martin-Serrano J. Caballe A, et al. Elife. 2015 May 26;4:e06547. doi: 10.7554/eLife.06547. Elife. 2015. PMID: 26011858 Free PMC article. - The Abscission Checkpoint: A Guardian of Chromosomal Stability.
Petsalaki E, Zachos G. Petsalaki E, et al. Cells. 2021 Nov 29;10(12):3350. doi: 10.3390/cells10123350. Cells. 2021. PMID: 34943860 Free PMC article. Review. - Knowing when to cut and run: mechanisms that control cytokinetic abscission.
Agromayor M, Martin-Serrano J. Agromayor M, et al. Trends Cell Biol. 2013 Sep;23(9):433-41. doi: 10.1016/j.tcb.2013.04.006. Epub 2013 May 22. Trends Cell Biol. 2013. PMID: 23706391 Review.
Cited by
- Midbody Remnant Inheritance Is Regulated by the ESCRT Subunit CHMP4C.
Casares-Arias J, González MU, San Paulo A, Ventimiglia LN, Sadler JBA, Miguez DG, Labat-de-Hoz L, Rubio-Ramos A, Rangel L, Bernabé-Rubio M, Fernández-Barrera J, Correas I, Martín-Serrano J, Alonso MA. Casares-Arias J, et al. iScience. 2020 Jun 26;23(6):101244. doi: 10.1016/j.isci.2020.101244. Epub 2020 Jun 7. iScience. 2020. PMID: 32629610 Free PMC article. - Aurora B kinase: a potential drug target for cancer therapy.
Ahmed A, Shamsi A, Mohammad T, Hasan GM, Islam A, Hassan MI. Ahmed A, et al. J Cancer Res Clin Oncol. 2021 Aug;147(8):2187-2198. doi: 10.1007/s00432-021-03669-5. Epub 2021 May 28. J Cancer Res Clin Oncol. 2021. PMID: 34047821 Review. - Spastin and ESCRT-III coordinate mitotic spindle disassembly and nuclear envelope sealing.
Vietri M, Schink KO, Campsteijn C, Wegner CS, Schultz SW, Christ L, Thoresen SB, Brech A, Raiborg C, Stenmark H. Vietri M, et al. Nature. 2015 Jun 11;522(7555):231-5. doi: 10.1038/nature14408. Epub 2015 Jun 3. Nature. 2015. PMID: 26040712 - Coordinated regulation of the ESCRT-III component CHMP4C by the chromosomal passenger complex and centralspindlin during cytokinesis.
Capalbo L, Mela I, Abad MA, Jeyaprakash AA, Edwardson JM, D'Avino PP. Capalbo L, et al. Open Biol. 2016 Oct;6(10):160248. doi: 10.1098/rsob.160248. Open Biol. 2016. PMID: 27784789 Free PMC article. - Cytokinetic abscission requires actin-dependent microtubule severing.
Advedissian T, Frémont S, Echard A. Advedissian T, et al. Nat Commun. 2024 Mar 2;15(1):1949. doi: 10.1038/s41467-024-46062-9. Nat Commun. 2024. PMID: 38431632 Free PMC article.
References
- Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316:1908–1912. - PubMed
- Caballe A, Martin-Serrano J. ESCRT machinery and cytokinesis: the road to daughter cell separation. Traffic. 2011;12:1318–1326. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 092429/Z/10/Z/WT_/Wellcome Trust/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- G0802777/MRC_/Medical Research Council/United Kingdom
- 093056/WT_/Wellcome Trust/United Kingdom
- WT093056MA/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous