MethLAB: a graphical user interface package for the analysis of array-based DNA methylation data - PubMed (original) (raw)
MethLAB: a graphical user interface package for the analysis of array-based DNA methylation data
Varun Kilaru et al. Epigenetics. 2012 Mar.
Abstract
Recent evidence suggests that DNA methylation changes may underlie numerous complex traits and diseases. The advent of commercial, array-based methods to interrogate DNA methylation has led to a profusion of epigenetic studies in the literature. Array-based methods, such as the popular Illumina GoldenGate and Infinium platforms, estimate the proportion of DNA methylated at single-base resolution for thousands of CpG sites across the genome. These arrays generate enormous amounts of data, but few software resources exist for efficient and flexible analysis of these data. We developed a software package called MethLAB (http://genetics.emory.edu/conneely/MethLAB) using R, an open source statistical language that can be edited to suit the needs of the user. MethLAB features a graphical user interface (GUI) with a menu-driven format designed to efficiently read in and manipulate array-based methylation data in a user-friendly manner. MethLAB tests for association between methylation and relevant phenotypes by fitting a separate linear model for each CpG site. These models can incorporate both continuous and categorical phenotypes and covariates, as well as fixed or random batch or chip effects. MethLAB accounts for multiple testing by controlling the false discovery rate (FDR) at a user-specified level. Standard output includes a spreadsheet-ready text file and an array of publication-quality figures. Considering the growing interest in and availability of DNA methylation data, there is a great need for user-friendly open source analytical tools. With MethLAB, we present a timely resource that will allow users with no programming experience to implement flexible and powerful analyses of DNA methylation data.
Figures
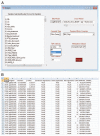
Figure 1
(A) A menu-driven Graphical User Interface (GUI) is used for loading methylation and phenotype files, selecting inclusion criteria and specifying the analytical model. (B) Analysis results are summarized in a spreadsheet-ready text file.
Figure 2
Manhattan plot of the negative log p values for each CpG site (vertical axis) by chromosome number and genomic position (horizontal axis). Dotted line indicates Holm significance.
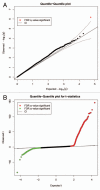
Figure 3
Automated quantile-quantile (Q-Q) plots based on (A) negative log p values from analysis of methylation and TLS and (B) ordered t statistics from analysis of sex-differential methylation.
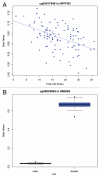
Figure 4
MethLAB automates (A) scatter or (B) box plots depicting methylation of specific CpG sites by continuous or categorical trait values. CpG probe and gene names are indicated in the plot titles.
Similar articles
- Efficient detection of differentially methylated regions using DiMmeR.
Almeida D, Skov I, Silva A, Vandin F, Tan Q, Röttger R, Baumbach J. Almeida D, et al. Bioinformatics. 2017 Feb 15;33(4):549-551. doi: 10.1093/bioinformatics/btw657. Bioinformatics. 2017. PMID: 27794558 - CpGassoc: an R function for analysis of DNA methylation microarray data.
Barfield RT, Kilaru V, Smith AK, Conneely KN. Barfield RT, et al. Bioinformatics. 2012 May 1;28(9):1280-1. doi: 10.1093/bioinformatics/bts124. Epub 2012 Mar 25. Bioinformatics. 2012. PMID: 22451269 Free PMC article. - An accessible GenePattern notebook for the copy number variation analysis of Illumina Infinium DNA methylation arrays.
Mah CK, Mesirov JP, Chavez L. Mah CK, et al. F1000Res. 2018 Dec 5;7:ISCB Comm J-1897. doi: 10.12688/f1000research.16338.1. eCollection 2018. F1000Res. 2018. PMID: 31105932 Free PMC article. - Base resolution methylome profiling: considerations in platform selection, data preprocessing and analysis.
Sun Z, Cunningham J, Slager S, Kocher JP. Sun Z, et al. Epigenomics. 2015 Aug;7(5):813-28. doi: 10.2217/epi.15.21. Epub 2015 Sep 14. Epigenomics. 2015. PMID: 26366945 Free PMC article. Review. - A comprehensive overview of Infinium HumanMethylation450 data processing.
Dedeurwaerder S, Defrance M, Bizet M, Calonne E, Bontempi G, Fuks F. Dedeurwaerder S, et al. Brief Bioinform. 2014 Nov;15(6):929-41. doi: 10.1093/bib/bbt054. Epub 2013 Aug 29. Brief Bioinform. 2014. PMID: 23990268 Free PMC article. Review.
Cited by
- Maternal adiposity negatively influences infant brain white matter development.
Ou X, Thakali KM, Shankar K, Andres A, Badger TM. Ou X, et al. Obesity (Silver Spring). 2015 May;23(5):1047-54. doi: 10.1002/oby.21055. Obesity (Silver Spring). 2015. PMID: 25919924 Free PMC article. - Higher levels of protective parenting are associated with better young adult health: exploration of mediation through epigenetic influences on pro-inflammatory processes.
Beach SR, Lei MK, Brody GH, Dogan MV, Philibert RA. Beach SR, et al. Front Psychol. 2015 May 28;6:676. doi: 10.3389/fpsyg.2015.00676. eCollection 2015. Front Psychol. 2015. PMID: 26074840 Free PMC article. - DNA methylation provides insight into intergenerational risk for preterm birth in African Americans.
Parets SE, Conneely KN, Kilaru V, Menon R, Smith AK. Parets SE, et al. Epigenetics. 2015;10(9):784-92. doi: 10.1080/15592294.2015.1062964. Epub 2015 Jun 19. Epigenetics. 2015. PMID: 26090903 Free PMC article. - Epigenomic association analysis identifies smoking-related DNA methylation sites in African Americans.
Sun YV, Smith AK, Conneely KN, Chang Q, Li W, Lazarus A, Smith JA, Almli LM, Binder EB, Klengel T, Cross D, Turner ST, Ressler KJ, Kardia SL. Sun YV, et al. Hum Genet. 2013 Sep;132(9):1027-37. doi: 10.1007/s00439-013-1311-6. Epub 2013 May 9. Hum Genet. 2013. PMID: 23657504 Free PMC article. - Analysing and interpreting DNA methylation data.
Bock C. Bock C. Nat Rev Genet. 2012 Oct;13(10):705-19. doi: 10.1038/nrg3273. Nat Rev Genet. 2012. PMID: 22986265 Review.
References
Publication types
MeSH terms
Grants and funding
- K01 MH085806/MH/NIMH NIH HHS/United States
- T32 ES007142/ES/NIEHS NIH HHS/United States
- MH088609/MH/NIMH NIH HHS/United States
- RC1 MH088609/MH/NIMH NIH HHS/United States
- MH085806/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources