Maternal siRNAs as regulators of parental genome imbalance and gene expression in endosperm of Arabidopsis seeds - PubMed (original) (raw)
Maternal siRNAs as regulators of parental genome imbalance and gene expression in endosperm of Arabidopsis seeds
Jie Lu et al. Proc Natl Acad Sci U S A. 2012.
Abstract
Seed size is important to crop domestication and natural selection and is affected by the balance of maternal and paternal genomes in endosperm. Endosperm, like placenta in mammals, provides reserves to the developing embryo. Interploidy crosses disrupt the genome balance in endosperm and alter seed size. Specifically, paternal-excess crosses (2 × 4) delay endosperm cellularization (EC) and produce larger seeds, whereas maternal-excess crosses (4 × 2) promote precocious EC and produce smaller seeds. The mechanisms for responding to the parental genome dosage imbalance and for gene expression changes in endosperm are unknown. In plants, RNA polymerase IV (PolIV or p4) encoded by NRPD1a is required for biogenesis of a major class of 24-nt small interfering RNAs (also known as p4-siRNAs), which are predominately expressed in developing endosperm. Here we show that p4-siRNA accumulation depends on the maternal genome dosage, and maternal p4-siRNAs target transposable elements (TEs) and TE-associated genes (TAGs) in seeds. The p4-siRNAs correlate negatively with expression levels of AGAMOUS-LIKE (AGL) genes in endosperm of interploidy crosses. Moreover, disruption of maternal NRPD1a expression is associated with p4-siRNA reduction and AGL up-regulation in endosperm of reciprocal crosses. This is unique genetic evidence for maternal siRNAs in response to parental genome imbalance and in control of transposons and gene expression during endosperm development.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
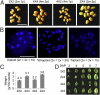
Seed morphology and chromosome counts in interploidy crosses. (A) Seed size and morphology in diploids, tetraploids, and triploids (2 × 4 or 4 × 2) in A. thaliana C24. m, maternal; p, paternal. By convention, the maternal parent is listed first in a genetic cross. (B) Chromosome counts in diploid, triploid, and tetraploid flowers in A. thaliana Col-0. (C) Seed weight in interploidy crosses. (D) Developing seeds dissected at 3–7 d after pollination (DAP) in Col-0.
Fig. 2.
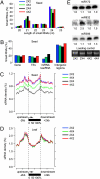
Small RNA distribution in interploidy crosses. (A) Size distribution of 20–25 nt small RNA reads in Col-0 seeds of 2 × 2 (blue), 4 × 4 (green), 2 × 4 (cyan), and 4 × 2 (magenta). (B) Distribution of 20–25 nt small RNAs in genes, TEs, miRNA, and ta-siRNA targets, and intergenic regions. (C and D) 24-nt small RNA densities (100-bp sliding window) in 5′ upstream (2 kb), transcribed, and 3′ downstream (2 kb) regions of TE genes in seeds (C) and leaves (D). (E) Small RNA blot analysis of miR172, miR832, and miR396 in diploids, triploids, and tetrpaloids at 6 DAP.
Fig. 3.
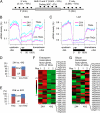
Distribution of TE-associated genes (_TAG_s) and parent-of-origin effects of siRNAs on TAG expression in reciprocal interploidy crosses. (A) Proportions of _TAG_s and locations of TEs (triangles) in coding sequences (gray box) and within 2 kb up or downstream of 5′ and 3′ regions (extended lines) in A. thaliana Col-0. (B and C) Small RNA densities (100-bp sliding window) in 5′ upstream (2 kb), transcribed, and 3′ downstream (2 kb) regions of _TAG_s and non-_TAG_s in seeds (B) and leaves (C). (D and E) Percentage of up-regulated (red) and down-regulated (blue) _TAG_s or non-_TAG_s in 2 × 4 vs. 4 × 2 crosses (D) and in 2 × 6 vs. 6 × 2 crosses (E). (F and G) Heat maps of gene expression changes in endosperm transcription factor (EFT) genes (n = 27) (F) and silique transcription factor (STF) genes (n = 25) (G) in two replicated experiments (Reps. 1 and 2); color bar indicates up (red) and down (green) regulation; black dots indicate up-regulated genes at statistically significant levels between 2 × 4 and 4 × 2 endosperm.
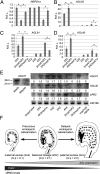
Fig. 4.
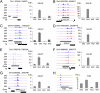
Maternal siRNAs are associated with expression of AGL genes and FWA. (A_–_H) siRNA hotspots (Left) and qRT-PCR analysis (Right) of AGL28 (A), AGL36 (B), AGL40 (C), AGL62 (D), AGL87 (E), AGL90 (F), AGL91 (G), and FWA (H) in 2 × 4 and 4 × 2 triploids and their parents (2 × 2 and 4 × 4). Diff., siRNA differences between 4 × 2 and 2 × 4; positive, Above the line; negative, Below the line; gray box, gene; black box, transposon. Genomic coordinates are shown Above each diagram, and SEs were calculated from three biological replicates.
Fig. 5.
siRNA production and AGL expression are dependent on RNA polymerase IV (NRPD1A) in endosperm and a model for endosperm development in interploidy crosses. (A) qRT-PCR analysis (relative expression levels, REL) of NRPD1a expression in endosperm of interploidy crosses. (B_–_D) qRT-PCR analyses of AGL62 (B), AGL91 (C), and AGL40 (D) expression (n = 3 biological replicates). (E) Small RNA blot analysis of p4-siRNA (siR02), _AGL40_-siRNA, and _AGL91_-siRNA in endosperm of interploidy crosses (n = 2 biological replicates). (Left) _AGL91_-siRNAs were present in seeds but not in siliques. miR166 was used as a control. (F) Model for the role of maternal siRNA-mediated AGL expression in endosperm and seed development (see text for explanation). Multiple dots and an elongated black rod in each diagram represent the endosperm and embryo cells, respectively.
Similar articles
- Paternally Acting Canonical RNA-Directed DNA Methylation Pathway Genes Sensitize Arabidopsis Endosperm to Paternal Genome Dosage.
Satyaki PRV, Gehring M. Satyaki PRV, et al. Plant Cell. 2019 Jul;31(7):1563-1578. doi: 10.1105/tpc.19.00047. Epub 2019 May 7. Plant Cell. 2019. PMID: 31064867 Free PMC article. - Maternal small RNAs mediate spatial-temporal regulation of gene expression, imprinting, and seed development in Arabidopsis.
Kirkbride RC, Lu J, Zhang C, Mosher RA, Baulcombe DC, Chen ZJ. Kirkbride RC, et al. Proc Natl Acad Sci U S A. 2019 Feb 12;116(7):2761-2766. doi: 10.1073/pnas.1807621116. Epub 2019 Jan 28. Proc Natl Acad Sci U S A. 2019. PMID: 30692258 Free PMC article. - Uniparental expression of PolIV-dependent siRNAs in developing endosperm of Arabidopsis.
Mosher RA, Melnyk CW, Kelly KA, Dunn RM, Studholme DJ, Baulcombe DC. Mosher RA, et al. Nature. 2009 Jul 9;460(7252):283-6. doi: 10.1038/nature08084. Epub 2009 Jun 3. Nature. 2009. PMID: 19494814 - Kin conflict in seed development: an interdependent but fractious collective.
Haig D. Haig D. Annu Rev Cell Dev Biol. 2013;29:189-211. doi: 10.1146/annurev-cellbio-101512-122324. Epub 2013 Apr 29. Annu Rev Cell Dev Biol. 2013. PMID: 23641801 Review. - Genomic imprinting and endosperm development in flowering plants.
Vinkenoog R, Bushell C, Spielman M, Adams S, Dickinson HG, Scott RJ. Vinkenoog R, et al. Mol Biotechnol. 2003 Oct;25(2):149-84. doi: 10.1385/MB:25:2:149. Mol Biotechnol. 2003. PMID: 14526125 Review.
Cited by
- Postzygotic reproductive isolation established in the endosperm: mechanisms, drivers and relevance.
Köhler C, Dziasek K, Del Toro-De León G. Köhler C, et al. Philos Trans R Soc Lond B Biol Sci. 2021 Jun 7;376(1826):20200118. doi: 10.1098/rstb.2020.0118. Epub 2021 Apr 19. Philos Trans R Soc Lond B Biol Sci. 2021. PMID: 33866810 Free PMC article. Review. - The MADS-box transcription factor PHERES1 controls imprinting in the endosperm by binding to domesticated transposons.
Batista RA, Moreno-Romero J, Qiu Y, van Boven J, Santos-González J, Figueiredo DD, Köhler C. Batista RA, et al. Elife. 2019 Dec 2;8:e50541. doi: 10.7554/eLife.50541. Elife. 2019. PMID: 31789592 Free PMC article. - Epigenetics Regulates Reproductive Development in Plants.
Han Q, Bartels A, Cheng X, Meyer A, An YC, Hsieh TF, Xiao W. Han Q, et al. Plants (Basel). 2019 Dec 2;8(12):564. doi: 10.3390/plants8120564. Plants (Basel). 2019. PMID: 31810261 Free PMC article. Review. - A Role for CHH Methylation in the Parent-of-Origin Effect on Altered Circadian Rhythms and Biomass Heterosis in Arabidopsis Intraspecific Hybrids.
Ng DW, Miller M, Yu HH, Huang TY, Kim ED, Lu J, Xie Q, McClung CR, Chen ZJ. Ng DW, et al. Plant Cell. 2014 Jun;26(6):2430-2440. doi: 10.1105/tpc.113.115980. Epub 2014 Jun 3. Plant Cell. 2014. PMID: 24894042 Free PMC article. - Genetic and molecular bases of yield-associated traits: a translational biology approach between rice and wheat.
Valluru R, Reynolds MP, Salse J. Valluru R, et al. Theor Appl Genet. 2014 Jul;127(7):1463-89. doi: 10.1007/s00122-014-2332-9. Epub 2014 Jun 10. Theor Appl Genet. 2014. PMID: 24913362 Review.
References
- Borlaug NE. Civilization's Future: A call for international granaries. Science and Public Affairs. 1973;24:7–15.
- Moore T, Haig D. Genomic imprinting in mammalian development: A parental tug-of-war. Trends Genet. 1991;7:45–49. - PubMed
- Stebbins GL. Seeds, seedlings, and the origin of angiosperms. In: Beck CB, editor. Origin and Early Evolution of Angiosperms. New York: Columbia Univ Press; 1976. pp. 300–311.
- Scott RJ, Spielman M, Bailey J, Dickinson HG. Parent-of-origin effects on seed development in Arabidopsis thaliana. Development. 1998;125:3329–3341. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials