Rac1 acts in conjunction with Nedd4 and dishevelled-1 to promote maturation of cell-cell contacts - PubMed (original) (raw)
. 2012 Jul 15;125(Pt 14):3430-42.
doi: 10.1242/jcs.100925. Epub 2012 Mar 30.
Bart-Jan de Kreuk, Daniele V F Tauriello, Eloise C Anthony, Barbara Snoek, Thomas Stumpel, Patricia C Salinas, Madelon M Maurice, Dirk Geerts, André M Deelder, Paul J Hensbergen, Peter L Hordijk
Affiliations
- PMID: 22467858
- PMCID: PMC3516380
- DOI: 10.1242/jcs.100925
Rac1 acts in conjunction with Nedd4 and dishevelled-1 to promote maturation of cell-cell contacts
Micha Nethe et al. J Cell Sci. 2012.
Abstract
The Rho-GTPase Rac1 promotes actin polymerization and membrane protrusion that mediate initial contact and subsequent maturation of cell-cell junctions. Here we report that Rac1 associates with the ubiquitin-protein ligase neural precursor cell expressed developmentally down-regulated 4 (Nedd4). This interaction requires the hypervariable C-terminal domain of Rac1 and the WW domains of Nedd4. Activated Rac1 colocalises with endogenous Nedd4 at epithelial cell-cell contacts. Reduction of Nedd4 expression by shRNA results in reduced transepithelial electrical resistance (TER) and concomitant changes in the distribution of adherens and tight junction markers. Conversely, expression of Nedd4 promotes TER, suggesting that Nedd4 cooperates with Rac1 in the induction of junctional maturation. We found that Nedd4, but not Nedd4-2, mediates the ubiquitylation and degradation of the adapter protein dishevelled-1 (Dvl1), the expression of which negatively regulates cell-cell contact. Nedd4-mediated ubiquitylation requires its binding to the C-terminal domain of Dvl1, comprising the DEP domain, and targets an N-terminal lysine-rich region upstream of the Dvl1 DIX domain. We found that endogenous Rac1 colocalises with endogenous Dvl1 in intracellular puncta as well as on cell-cell junctions. Finally, activated Rac1 was found to stimulate Nedd4 activity, resulting in increased ubiquitylation of Dvl1. Together, these data reveal a novel Rac1-dependent signalling pathway that, through Nedd4-mediated ubiquitylation of Dvl1, stimulates the maturation of epithelial cell-cell contacts.
Figures
Fig. 1.
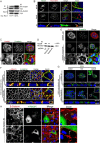
Nedd4 associates with Rac1. (A) Silver stained SDS-PAGE gel showing proteins binding to the Rac1 C-terminal peptide. The protein identified by mass spectrometry as Nedd4 is indicated (arrow). (B) Biotinylated peptides encoding the C-terminal domains of different RhoGTPases were assayed for their ability to bind endogenous Nedd4. (C) The interaction of Rac1 with Nedd4 was further examined using different peptides resembling part of the Rac1 effector domain (17–32) and the Rac1 hypervariable C-terminal region (Rac1) or mutated versions thereof, in which the three prolines (Rac1 P-A) or the polybasic region (Rac1 RKR-AAA) were mutated to alanines. Binding of the peptides to the Rac1 GEF β-PIX was included as a control. (D) GST-tagged Rac1 and Rac1ΔC, which lacks the C-terminal region, were examined using a pull-down (pd) assay of HeLa cell lysates for binding to HA-tagged Nedd4. EV, empty vector. (E) HeLa cells were transfected with WT Nedd4 or the Nedd4ΔC2 mutant and tested for differential binding to the Rac1 or Rac2 C-terminal peptides. Binding to PACSIN2 (de Kreuk et al., 2011) was included as a control. (F) Binding of GST–Rac1 to GFP fusions of Nedd4 WT or of the WW-domain region (Nedd4 WW1-4) in HeLa cell lysates.
Fig. 2.
Nedd4 promotes maturation of cell–cell junctions. (A) Immunoprecipitated, myc-tagged inactive (N17T) and active (Q61L) Rac1 were analysed for binding to co-expressed HA–Nedd4 in HeLa cells. (B) HeLa cells, transfected with myc–Rac1Q61L, were fixed and immunostained for endogenous Nedd4 and myc. Z-stacks along the dashed line were analysed for colocalisation of Nedd4 with active Rac1. Arrows indicate colocalisation of Nedd4 with active Rac1 at cell–cell junctions (left arrow in XZ image) and membrane ruffles. Scale bar: 10 µm. (C) HeLa cells were fixed and immunostained for endogenous Nedd4 and β-catenin. Arrows indicate the absence of Nedd4 at immature junctions (left in ‘zoom’ images) and its presence at mature junctions. Scale bar: 20 µm. (D) Western blot showing the reduction of endogenous Nedd4 protein upon transduction of HeLa cells with lentiviral shRNAs. (E) Confocal imaging for Nedd4 and β-catenin in HeLa cells transduced with the control or Nedd4 shRNA. Scale bar: 20 µm. (F) H292 lung epithelial cells were fixed 48 hrs after transfection with the indicated shRNAs and immunostained for endogenous β-catenin and γ-catenin. XZ images were derived from Z-stack sections along the dashed lines. Scale bar: 15 µm. (G) As for F, H292 cells were stained for endogenous ZO-1 as a marker of tight junctions (arrows in XZ image). XZ images were derived from Z-stack sections along the dashed lines. Scale bar: 15 µm. Boxed areas are magnified in the insets. (H) HeLa cells were transduced with control or Nedd4 shRNA and subsequently transfected with murine HA–Nedd4. Cells were analysed for distribution of β-catenin and murine Nedd4 expression. Two representative images are shown; arrows indicate mature, linear cell–cell contacts. Scale bars: 10 µm.
Fig. 3.
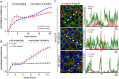
Nedd4 increases TER and associates with mature cell–cell contacts. (A) Cell spreading and monolayer formation of control and Nedd4-shRNA-transduced H292 cells were analysed by ECIS and plotted as normalised resistance (n = 4; values are means ± s.e.m.). (B) ECIS analysis of cell spreading and monolayer formation of GFP and GFP–Nedd4-expressing, FACS-sorted HeLa cells. Data are plotted as normalised resistance (n = 4; values are means ± s.e.m.). Note the difference in resistance between A and B, which is due to the use of different cell types. (C) Confocal images of H292 cells immunostained for endogenous Nedd4 and β-catenin following disruption of junctions with EGTA (upper panel) and reformation of cell–cell contacts (1 hr and 5 hr washout), in a calcium-switch assay. Profile scan along the dashed lines shows loss and restoration of Nedd4 colocalisation with β-catenin.
Fig. 4.
Nedd4 regulates ubiquitylation of Dvl1. (A) Schematic representation of Dvl1 domains showing the structure and position of the PPXY motif. (B) Calcium-switch assay of H292 cells expressing Dvl1–GFP. Reconstitution of cell–cell junctions was analysed by immunostaining for β-catenin. Profile scan depicts the distribution of β-catenin along the dashed lines in control and Dvl1–GFP-expressing cells. Matched arrows in images and profile scan indicate cell–cell contacts. Scale bar: 10 µm. (C) Protein expression levels of endogenous Dvl1, -2 and -3 were assessed in control and GFP–Nedd4 expressing HeLa cells, treated for 6 hrs with Cycloheximide (CHX; 10 µg/ml). Bar graph includes quantification of Dvl1 protein levels (n = 3; mean ± s.e.m.). α-catenin and RhoGDI were included as loading controls. (D) Stability of Dvl proteins in H292 cells 48 hrs after transfection with the indicated shRNAs and 6 hr after incubation with CHX (10 µg/ml). (E) Schematic of the domain structure of Nedd4 and Nedd4-2. Ubiquitylation of Dvl1–HA was assessed in empty vector (GFP-EV), GFP–Nedd4 or GFP–Nedd4-2 co-transfected HeLa cells, that were treated for 6 hrs with MG132 (25 µM) or chloroquine (100 µM) to inhibit lysosomal degradation prior to cell lysis and analysis of ubiquitylation.
Fig. 5.
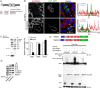
Mapping of the Nedd4–Dvl1 interaction and ubiquitylation. (A) Nedd4 fragments fused to GFP encoding the N-terminus (Nedd4-C2) and the middle region including the four WW domains (Nedd4-WW) were expressed in HeLa cells and tested for binding to GST (EV) or GST–Dvl1. (B) Overview of the different Dvl1 truncation and deletion mutants is shown in the top panel. Lower panels show analysis of the binding of FLAG–Dvl1 constructs, expressed in HeLa cells, to GST (EV) or GST–Nedd4. (C) Nedd4-driven ubiquitylation of Dvl1 was analysed using the indicated Dvl1–HA deletion mutants. (D) The requirement of the Dvl1 C-terminal region including the DEP domain, for Nedd4-induced ubiquitylation was examined using the indicated FLAG-tagged Dvl1 constructs. (E) Nedd4-driven ubiquitylation of HA-tagged Dvl1 was compared with that of the indicated PPXY motif mutant of Dvl1 (P551A/Y553F). (F) Mutation of lysine residues K218, K220, K225 to arginine (3KR), deletion of the DEP domain and adjacent C-terminal portion as well as deletion of the N-terminal portion up to the PDZ domain all impaired Nedd4-mediated reduction of Dvl1 protein levels, indicative for reduced ubiquitylation and degradation. (G) Expression of Dvl1 WT and the Dvl1 3KR protein reduced TER, as measured by ECIS in HeLa cells, seeded on gold electrodes. The arrow indicates the time point of the quantification in the bar graph. (H) Schematic overview summarising the data on the Nedd4–Dvl1 interaction and ubiquitylation of Dvl1, indicating that the binding to Nedd4 is mediated by the Dvl1 DEP domain and PPXY motif, and that ubiquitylation and degradation require the three lysines in between the DIX and PDZ domains. *P<0.05, **P<0.01.
Fig. 6.
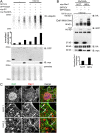
Dvl1 ubiquitylation and junctional localisation of Nedd4 require the C2 domain. (A) GFP–Nedd4 colocalises with Dvl1–HA in puncta and in part with β-catenin at cell–cell junctions in H292 cells (arrows). Scale bar: 10 µm. (B) Endogenous Dvl1 colocalises with GFP–Nedd4 in puncta as well as along cell–cell contacts (arrows). Scale bar: 10 µm. (C) Subcellular distribution of GFP–ΔC2-Nedd4 in H292 cells reveals its absence at cell-cell contacts (arrows). The XZ images are derived from a section corresponding with the dashed line. Scale bar, 20 µm. In A–C the boxed regions are enlarged in the lower panels (zoom). (D) Ubiquitylation of Dvl1 by Nedd4 lacking the C2 domain (GFP–ΔC2-Nedd4) was lost compared with its ubiquitylation by full-length Nedd4. The catalytically inactive GFP–Nedd4C867S was included as a control.
Fig. 7.
Activated Rac1 promotes Nedd4-mediated ubiquitylation. (A) Analysis of Rac1-stimulated, Nedd4-mediated protein ubiquitylation in cell lysates, blotted for ubiquitin. The bar graph shows quantification of the overall ubiquitin signal. kd, kilodaltons. (B) Stimulation by activated (Q61L)Rac1 compared with (N17T)Rac1 of Nedd4-mediated ubiquitylation of Dvl1–HA, co-transfected with His–ubiquitin. kD, kilodaltons. (C) Immunostaining to show colocalisation of endogenous Dvl1 and endogenous Rac1 in puncta (zoom 1, arrows) as well as along cell–cell contacts (zoom 2, arrow). Scale bar: 5 µm.
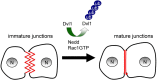
Fig. 8.
Model for regulation of cell–cell contacts by the Nedd4-mediated ubiquitylation of Dvl1. In nascent cell–cell contacts, Rac1 activity recruits Nedd4 to intercellular junctions. Nedd4, stimulated by activated Rac1, regulates the poly-ubiquitylation and degradation of Dvl1. The local loss of Dvl1 promotes the maturation of cell–cell contacts and the formation of strong intercellular adhesion.
Similar articles
- Ubiquitylation of Fe65 adaptor protein by neuronal precursor cell expressed developmentally down regulated 4-2 (Nedd4-2) via the WW domain interaction with Fe65.
Lee EJ, Hyun S, Chun J, Shin SH, Kang SS. Lee EJ, et al. Exp Mol Med. 2009 Aug 31;41(8):555-68. doi: 10.3858/emm.2009.41.8.061. Exp Mol Med. 2009. PMID: 19381069 Free PMC article. - HECT domain-containing E3 ubiquitin ligase NEDD4L negatively regulates Wnt signaling by targeting dishevelled for proteasomal degradation.
Ding Y, Zhang Y, Xu C, Tao QH, Chen YG. Ding Y, et al. J Biol Chem. 2013 Mar 22;288(12):8289-8298. doi: 10.1074/jbc.M112.433185. Epub 2013 Feb 9. J Biol Chem. 2013. PMID: 23396981 Free PMC article. - YAP1 recruits c-Abl to protect angiomotin-like 1 from Nedd4-mediated degradation.
Skouloudaki K, Walz G. Skouloudaki K, et al. PLoS One. 2012;7(4):e35735. doi: 10.1371/journal.pone.0035735. Epub 2012 Apr 27. PLoS One. 2012. PMID: 22558212 Free PMC article. - Nedd4 and Nedd4-2: closely related ubiquitin-protein ligases with distinct physiological functions.
Yang B, Kumar S. Yang B, et al. Cell Death Differ. 2010 Jan;17(1):68-77. doi: 10.1038/cdd.2009.84. Cell Death Differ. 2010. PMID: 19557014 Free PMC article. Review. - Regulation of functional diversity within the Nedd4 family by accessory and adaptor proteins.
Shearwin-Whyatt L, Dalton HE, Foot N, Kumar S. Shearwin-Whyatt L, et al. Bioessays. 2006 Jun;28(6):617-28. doi: 10.1002/bies.20422. Bioessays. 2006. PMID: 16700065 Review.
Cited by
- The N and C termini of ZO-1 are surrounded by distinct proteins and functional protein networks.
Van Itallie CM, Aponte A, Tietgens AJ, Gucek M, Fredriksson K, Anderson JM. Van Itallie CM, et al. J Biol Chem. 2013 May 10;288(19):13775-88. doi: 10.1074/jbc.M113.466193. Epub 2013 Apr 3. J Biol Chem. 2013. PMID: 23553632 Free PMC article. - Rac1 Signaling: From Intestinal Homeostasis to Colorectal Cancer Metastasis.
Kotelevets L, Chastre E. Kotelevets L, et al. Cancers (Basel). 2020 Mar 12;12(3):665. doi: 10.3390/cancers12030665. Cancers (Basel). 2020. PMID: 32178475 Free PMC article. Review. - Peptide and small molecule inhibitors of HECT-type ubiquitin ligases.
Mund T, Lewis MJ, Maslen S, Pelham HR. Mund T, et al. Proc Natl Acad Sci U S A. 2014 Nov 25;111(47):16736-41. doi: 10.1073/pnas.1412152111. Epub 2014 Nov 10. Proc Natl Acad Sci U S A. 2014. PMID: 25385595 Free PMC article. - Proteomic analysis of proteins surrounding occludin and claudin-4 reveals their proximity to signaling and trafficking networks.
Fredriksson K, Van Itallie CM, Aponte A, Gucek M, Tietgens AJ, Anderson JM. Fredriksson K, et al. PLoS One. 2015 Mar 19;10(3):e0117074. doi: 10.1371/journal.pone.0117074. eCollection 2015. PLoS One. 2015. PMID: 25789658 Free PMC article. - The Daam2-VHL-Nedd4 axis governs developmental and regenerative oligodendrocyte differentiation.
Ding X, Jo J, Wang CY, Cristobal CD, Zuo Z, Ye Q, Wirianto M, Lindeke-Myers A, Choi JM, Mohila CA, Kawabe H, Jung SY, Bellen HJ, Yoo SH, Lee HK. Ding X, et al. Genes Dev. 2020 Sep 1;34(17-18):1177-1189. doi: 10.1101/gad.338046.120. Epub 2020 Aug 13. Genes Dev. 2020. PMID: 32792353 Free PMC article.
References
- Boyer L., Turchi L., Desnues B., Doye A., Ponzio G., Mege J. L., Yamashita M., Zhang Y. E., Bertoglio J., Flatau G., et al. (2006). CNF1-induced ubiquitylation and proteasome destruction of activated RhoA is impaired in Smurf1-/- cells. Mol. Biol. Cell 17, 2489–2497 10.1091/mbc.E05-09-0876 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials