CRISPR typing and subtyping for improved laboratory surveillance of Salmonella infections - PubMed (original) (raw)
doi: 10.1371/journal.pone.0036995. Epub 2012 May 18.
Jian Zhang, Ghislaine Guigon, Simon Le Hello, Véronique Guibert, Marie Accou-Demartin, Saïana de Romans, Catherine Lim, Chrystelle Roux, Virginie Passet, Laure Diancourt, Martine Guibourdenche, Sylvie Issenhuth-Jeanjean, Mark Achtman, Sylvain Brisse, Christophe Sola, François-Xavier Weill
Affiliations
- PMID: 22623967
- PMCID: PMC3356390
- DOI: 10.1371/journal.pone.0036995
CRISPR typing and subtyping for improved laboratory surveillance of Salmonella infections
Laëtitia Fabre et al. PLoS One. 2012.
Abstract
Laboratory surveillance systems for salmonellosis should ideally be based on the rapid serotyping and subtyping of isolates. However, current typing methods are limited in both speed and precision. Using 783 strains and isolates belonging to 130 serotypes, we show here that a new family of DNA repeats named CRISPR (clustered regularly interspaced short palindromic repeats) is highly polymorphic in Salmonella. We found that CRISPR polymorphism was strongly correlated with both serotype and multilocus sequence type. Furthermore, spacer microevolution discriminated between subtypes within prevalent serotypes, making it possible to carry out typing and subtyping in a single step. We developed a high-throughput subtyping assay for the most prevalent serotype, Typhimurium. An open web-accessible database was set up, providing a serotype/spacer dictionary and an international tool for strain tracking based on this innovative, powerful typing and subtyping tool.
Conflict of interest statement
Competing Interests: The authors have read the journal’s policy and have the following conflicts: Some of the authors (FXW, LF, VG, LD, and SB) have filed an international patent application (no. PCT/IB2008/004004) for the molecular typing and subtyping of Salmonella by identification of the variable nucleotide sequences of the CRISPR loci. This does not alter the authors’ adherence to all the PLoS ONE policies on sharing data and materials.
Figures
Figure 1. CRISPR/Cas system structures from 39 available genome sequences for S. enterica and S. bongori.
Two CRISPR loci (CRISPR1 and CRISPR2) are present in all genomes. The CRISPR-associated (cas) genes cas2, cas1, cse3, cas5e, cse4, cse2, and cas3 genes of the “Ecoli” subtype are located between the CRISPR loci. The most frequent structure, A, is represented by S. enterica serotype Typhimurium strain LT2. Structures B to E are represented by S. enterica serotypes Choleraesuis strain SC-B67, Javiana strain GA_MM04042433, Paratyphi B strain SPB7, and S. enterica subsp. arizonae serotype 62:z4,z23:- strain CDC346-86, respectively. Structure F is represented by S. enterica serotype Typhi strain Ty2. Black diamonds represent direct repeats, with colored diamonds indicating spacers. The CRISPR1 locus of serotype Typhi strain Ty2 is enlarged. The primers used to amplify and sequence the CRISPR loci for the spacer inventory are indicated by horizontal arrows.
Figure 2. Multilocus sequence typing and CRISPR spacer content of 34 S. enterica strains and isolates with the antigenic formula 6,7:c:1,5.
Based on additional biochemical characters, we can identify five subserotypes: Choleraesuis sensu stricto, Choleraesuis variant Kunzendorf, Paratyphi C (human-restricted), Typhisuis (pig-restricted) and Decatur. MLST (a) and CRISPR data (b) show that Choleraesuis, Paratyphi C and Typhisuis share a common ancestor, whereas Decatur is made up of at least five unrelated populations. The numbers in panel “a” correspond to the allelic difference between STs. The size of the circle is not correlated with the number of strains with the corresponding ST. The exact name of the spacers and the spacer content of Decatur strains from panel b can be found in Table S2.
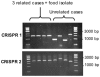
Figure 3. CRISPR sizing by PCR for the rapid comparison of Salmonella spp isolates.
Results of PCR amplification for 8 S. enterica serotype Typhimurium isolates collected from the same city during a single week (cluster E in Table S7). Three cases were from the same food poisoning cluster (the food isolate was also tested), whereas the other cases were unrelated.
Figure 4. Overview of the bead-based CRISPOL assay for S. enterica serotype Typhimurium developed here.
The estimated time for each step is based on the testing of 65 isolates in a 96-well plate. The amplification step spans from the preparation of thermolysates to the gel electrophoresis of PCR products.
Figure 5. Dendrogram presentation of the 245 distinct CRISPOL types detected among 2,200 isolates of S. enterica serotype Typhimurium or its monophasic variant of antigenic formula 1,4,,12:i:-.
Black squares indicate presence of the spacer, as detected by the corresponding probe, whereas white indicates an absence of the spacer. For the determination of CRISPOL types (CTs), each of the 68 spacers was treated as a numerical character indicating absence (0) or presence (1 for all spacers except BraB14, for which an arbitrary value of 10 was assigned) in BioNumerics 6.5 software (Applied Maths, Sint-Martens-Latem, Belgium). Similarities between CTs were assessed by calculating the Pearson product-moment, and a dendrogram was constructed by the unweighted pair group method with arithmetic mean (UPGMA). The four SNP-variant spacers targeted by probes 69 to 72 are shown but were excluded from the phylogenetic analysis, as they were not independent. A indicates a group of profiles derived from CT1, the main type of emerging monophasic isolates. B indicates a group of profiles derived from CT21, which is associated with multidrug-resistant DT104 serotype Typhimurium isolates. C indicates a group of serotype Typhimurium isolates of ST36 that may have one or two specific spacers on the leader side of CRISPR1 (BraB14) and CRISPR2 (STMB35).
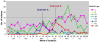
Figure 6. Distribution of selected CRISPOL types of S. enterica serotype Typhimurium and S. enterica with antigenic formula 1,4,,12:i :- isolated from humans in France between January 1 and July 4 2010.
Over this period, all 1,084 isolates (one per patient) were CRISPOL-typed and two outbreaks were investigated. Outbreak A (≈40 cases) was due to the consumption of a raw milk cheese contaminated with a CT62 highly multidrug-resistant S. enterica serotype Typhimurium strain, whereas outbreak B (≈50 cases) was caused by the consumption of a dried pork sausage contaminated with a CT136 S. enterica 4,12:i:- strain. The third peak, corresponding to CT21, in May was neither detected nor investigated, as CRISPOL typing was carried out retrospectively.
Similar articles
- New clustered regularly interspaced short palindromic repeat locus spacer pair typing method based on the newly incorporated spacer for Salmonella enterica.
Li H, Li P, Xie J, Yi S, Yang C, Wang J, Sun J, Liu N, Wang X, Wu Z, Wang L, Hao R, Wang Y, Jia L, Li K, Qiu S, Song H. Li H, et al. J Clin Microbiol. 2014 Aug;52(8):2955-62. doi: 10.1128/JCM.00696-14. Epub 2014 Jun 4. J Clin Microbiol. 2014. PMID: 24899040 Free PMC article. - Use of molecular subtyping in surveillance for Salmonella enterica serotype typhimurium.
Bender JB, Hedberg CW, Boxrud DJ, Besser JM, Wicklund JH, Smith KE, Osterholm MT. Bender JB, et al. N Engl J Med. 2001 Jan 18;344(3):189-95. doi: 10.1056/NEJM200101183440305. N Engl J Med. 2001. PMID: 11172141 - Novel virulence gene and clustered regularly interspaced short palindromic repeat (CRISPR) multilocus sequence typing scheme for subtyping of the major serovars of Salmonella enterica subsp. enterica.
Liu F, Barrangou R, Gerner-Smidt P, Ribot EM, Knabel SJ, Dudley EG. Liu F, et al. Appl Environ Microbiol. 2011 Mar;77(6):1946-56. doi: 10.1128/AEM.02625-10. Epub 2011 Jan 28. Appl Environ Microbiol. 2011. PMID: 21278266 Free PMC article. - Molecular methods for serovar determination of Salmonella.
Shi C, Singh P, Ranieri ML, Wiedmann M, Moreno Switt AI. Shi C, et al. Crit Rev Microbiol. 2015;41(3):309-25. doi: 10.3109/1040841X.2013.837862. Epub 2013 Nov 14. Crit Rev Microbiol. 2015. PMID: 24228625 Review. - CRISPRs: molecular signatures used for pathogen subtyping.
Shariat N, Dudley EG. Shariat N, et al. Appl Environ Microbiol. 2014 Jan;80(2):430-9. doi: 10.1128/AEM.02790-13. Epub 2013 Oct 25. Appl Environ Microbiol. 2014. PMID: 24162568 Free PMC article. Review.
Cited by
- Evidence for the widespread distribution of CRISPR-Cas system in the Phylum Cyanobacteria.
Cai F, Axen SD, Kerfeld CA. Cai F, et al. RNA Biol. 2013 May;10(5):687-93. doi: 10.4161/rna.24571. Epub 2013 Apr 11. RNA Biol. 2013. PMID: 23628889 Free PMC article. - CRISPR-Cas: evolution of an RNA-based adaptive immunity system in prokaryotes.
Koonin EV, Makarova KS. Koonin EV, et al. RNA Biol. 2013 May;10(5):679-86. doi: 10.4161/rna.24022. Epub 2013 Feb 25. RNA Biol. 2013. PMID: 23439366 Free PMC article. Review. - Study the Features of 57 Confirmed CRISPR Loci in 38 Strains of Staphylococcus aureus.
Zhao X, Yu Z, Xu Z. Zhao X, et al. Front Microbiol. 2018 Jul 26;9:1591. doi: 10.3389/fmicb.2018.01591. eCollection 2018. Front Microbiol. 2018. PMID: 30093886 Free PMC article. - Fourier transform infrared spectroscopy: unlocking fundamentals and prospects for bacterial strain typing.
Novais Â, Freitas AR, Rodrigues C, Peixe L. Novais Â, et al. Eur J Clin Microbiol Infect Dis. 2019 Mar;38(3):427-448. doi: 10.1007/s10096-018-3431-3. Epub 2018 Nov 27. Eur J Clin Microbiol Infect Dis. 2019. PMID: 30483997 Review. - Distribution and genomic characterization of tigecycline-resistant tet(X4)-positive Escherichia coli of swine farm origin.
Li Y, Wang Q, Peng K, Liu Y, Xiao X, Mohsin M, Li R, Wang Z. Li Y, et al. Microb Genom. 2021 Oct;7(10):000667. doi: 10.1099/mgen.0.000667. Microb Genom. 2021. PMID: 34693904 Free PMC article.
References
- Voetsch AC, Van Gilder TJ, Angulo FJ, Farley MM, Shallow S, et al. FoodNet estimate of the burden of illness caused by nontyphoidal Salmonella infections in the United States. Clin Infect. 2004;Dis(Suppl 3):S127–134. - PubMed
- Fisher IS, Enter-net participants. International trends in Salmonella serotypes 1998–2003–a surveillance report from the Enter-net international surveillance network. Euro Surveill. 2004;9:45–47. - PubMed
- Grimont PAD, Weill FX. Antigenic formulae of the Salmonella serovars. 9th ed. Paris, France: WHO Collaborating Center for Reference and Research on Salmonella, Institut Pasteur website. 2007;23 Available : http://www.pasteur.fr/ip/portal/action/WebdriveActionEvent/oid/01s-00003.... Accessed 2012 Apr.
- Bender JB, Hedberg CW, Boxrud DJ, Besser JM, Wicklund JH, et al. Use of molecular subtyping in surveillance for Salmonella enterica serotype Typhimurium. N Engl J Med. 2001;344:189–195. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases