K63-linked ubiquitination in kinase activation and cancer - PubMed (original) (raw)
K63-linked ubiquitination in kinase activation and cancer
Guocan Wang et al. Front Oncol. 2012.
Abstract
Ubiquitination has been demonstrated to play a pivotal role in multiple biological functions, which include cell growth, proliferation, apoptosis, DNA damage response, innate immune response, and neuronal degeneration. Although the role of ubiquitination in targeting proteins for proteasome-dependent degradation have been extensively studied and well-characterized, the critical non-proteolytic functions of ubiquitination, such as protein trafficking and kinase activation, involved in cell survival and cancer development, just start to emerge, In this review, we will summarize recent progresses in elucidating the non-proteolytic function of ubiquitination signaling in protein kinase activation and its implications in human cancers. The advancement in the understanding of the novel functions of ubiquitination in signal transduction pathways downstream of growth factor receptors may provide novel paradigms for the treatment of human cancers.
Keywords: Akt; NF-κB; TRAF6; phosphorylation; protein kinase; tumorigenesis; ubiquitination.
Figures
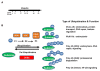
Figure 1
(A) Ubiquitin, a 76-amino acid protein, is highly evolutionarily conserved across species. It contains seven lysine (K) residues (K6, K11, K27, K29, K33, K48, and K63) in the ubiquitin, which have been identified to be utilized for ubiquitination chains. (B) In the ubiquitination process, ubiquitin is covalently conjugated to lysine residues of substrate proteins through a three-step enzymatic reaction. Ubiquitin is first activated by ubiquitin-activating enzyme (E1). The activated ubiquitin is then delivered to the ubiquitin-conjugating enzyme (E2), which will be transferred from the E2 to a lysine residue of a target protein by ubiquitin ligase (E3). The function usefulness of ubiquitin conjugation is not limited to the ubiquitin–proteasome pathway. Mono- and polyubiquitins are used as signals in various pathways including endocytosis, DNA repair, apoptosis, and transcriptional regulation.
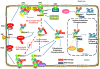
Figure 2
The role of K63-linked polyubiquitination in the activation of canonical and non-canonical NF-κB signaling pathway. In both canonical and non-canonical NF-κB pathways, ligand binding to receptors results in the recruitment of the TNF receptor-associated factors (TRAFs) to the receptors. In the canonical pathway, the recruitment of E3 ligase TRAF6promotesits auto-polyubiquitinating and other neighboring target proteins via K63-linked polyubiquitin chains. Unanchored free K63-linked polyubiquitin then directly activate TAK1 kinase, which then phosphorylates and activates IKK-β to induce NF- κB activation. In the non-canonical pathway, TRAF3, TRAF2, and cIAP promote rapid proteasomal degradation of NIK by the ubiquitin–proteasome system under un-stimulated condition. Various stimuli release NIK from TRAF3 inhibition by triggering cIAP activation by TRAF2-mediated K63-polyubiquitination and subsequent degradation of TRAF3. NIK-mediated-phosphorylation of p100 and its subsequent processing promotes RelB/p52 nuclear translocation and NF- κB activation.
Figure 3
Ubiquitination regulates Akt stability and activation. IGF-1-induced activation of IGF-1R promotes TRAF6 activation, which then triggers K63-linked ubiquitination of Akt and promotes Akt membrane recruitment and subsequent phosphorylation by PDK1 and mTORC2. Active Akt then phosphorylates its substrates in the cytoplasm and nucleus to exert its biological functions. E3 ligase TTC3 has been suggested to play a role in the termination of active Akt signaling in the nucleus by promoting Akt ubiquitination and degradation by the proteasome. TTC3 activity is regulated by Akt-mediated phosphorylation at S378, thus providing a negative feedback loop for Akt inactivation. In addition, Akt ubiquitination is promoted by CHIP or BRCA1 and leads to Akt degradation.
Similar articles
- Emerging role of Lys-63 ubiquitination in protein kinase and phosphatase activation and cancer development.
Yang WL, Zhang X, Lin HK. Yang WL, et al. Oncogene. 2010 Aug 12;29(32):4493-503. doi: 10.1038/onc.2010.190. Epub 2010 Jun 7. Oncogene. 2010. PMID: 20531303 Free PMC article. Review. - The role of K63-linked polyubiquitination in cardiac hypertrophy.
Yan K, Ponnusamy M, Xin Y, Wang Q, Li P, Wang K. Yan K, et al. J Cell Mol Med. 2018 Oct;22(10):4558-4567. doi: 10.1111/jcmm.13669. Epub 2018 Aug 13. J Cell Mol Med. 2018. PMID: 30102008 Free PMC article. Review. - Elucidating dynamic protein-protein interactions and ubiquitination in NF-κB signaling pathways.
Shembade N, Harhaj EW. Shembade N, et al. Methods Mol Biol. 2015;1280:283-95. doi: 10.1007/978-1-4939-2422-6_16. Methods Mol Biol. 2015. PMID: 25736755 - Hepatitis B e Antigen Inhibits NF-κB Activity by Interrupting K63-Linked Ubiquitination of NEMO.
Wang Y, Cui L, Yang G, Zhan J, Guo L, Chen Y, Fan C, Liu D, Guo D. Wang Y, et al. J Virol. 2019 Jan 4;93(2):e00667-18. doi: 10.1128/JVI.00667-18. Print 2019 Jan 15. J Virol. 2019. PMID: 30404796 Free PMC article. - pVHL mediates K63-linked ubiquitination of IKKβ, leading to IKKβ inactivation.
Wang Y, Zhao W, Gao Q, Fan L, Qin Y, Zhou H, Li M, Fang J. Wang Y, et al. Cancer Lett. 2016 Dec 1;383(1):1-8. doi: 10.1016/j.canlet.2016.09.009. Epub 2016 Sep 28. Cancer Lett. 2016. PMID: 27693634
Cited by
- The diversity of linkage-specific polyubiquitin chains and their role in synaptic plasticity and memory formation.
Musaus M, Navabpour S, Jarome TJ. Musaus M, et al. Neurobiol Learn Mem. 2020 Oct;174:107286. doi: 10.1016/j.nlm.2020.107286. Epub 2020 Aug 1. Neurobiol Learn Mem. 2020. PMID: 32745599 Free PMC article. Review. - The Role of Deubiquitinating Enzyme in Head and Neck Squamous Cell Carcinoma.
Jin S, Kudo Y, Horiguchi T. Jin S, et al. Int J Mol Sci. 2022 Dec 29;24(1):552. doi: 10.3390/ijms24010552. Int J Mol Sci. 2022. PMID: 36613989 Free PMC article. Review. - Non-proteolytic ubiquitylation in cellular signaling and human disease.
Liao Y, Sumara I, Pangou E. Liao Y, et al. Commun Biol. 2022 Feb 8;5(1):114. doi: 10.1038/s42003-022-03060-1. Commun Biol. 2022. PMID: 35136173 Free PMC article. Review. - USP10 modulates the SKP2/Bcr-Abl axis via stabilizing SKP2 in chronic myeloid leukemia.
Liao Y, Liu N, Xia X, Guo Z, Li Y, Jiang L, Zhou R, Tang D, Huang H, Liu J. Liao Y, et al. Cell Discov. 2019 Apr 30;5:24. doi: 10.1038/s41421-019-0092-z. eCollection 2019. Cell Discov. 2019. PMID: 31044085 Free PMC article. - TRIM24 facilitates antiviral immunity through mediating K63-linked TRAF3 ubiquitination.
Zhu Q, Yu T, Gan S, Wang Y, Pei Y, Zhao Q, Pei S, Hao S, Yuan J, Xu J, Hou F, Wu X, Peng C, Wu P, Qin J, Xiao Y. Zhu Q, et al. J Exp Med. 2020 Jul 6;217(7):e20192083. doi: 10.1084/jem.20192083. J Exp Med. 2020. PMID: 32324863 Free PMC article.
References
- Adhikary S., Marinoni F., Hock A., Hulleman E., Popov N., Beier R., Bernard S., Quarto M., Capra M., Goettig S., Kogel U., Scheffner M., Helin K., Eilers M. (2005). The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell 123, 409–42110.1016/j.cell.2005.08.016 - DOI - PubMed
LinkOut - more resources
Full Text Sources