RecQ helicase translocates along single-stranded DNA with a moderate processivity and tight mechanochemical coupling - PubMed (original) (raw)
RecQ helicase translocates along single-stranded DNA with a moderate processivity and tight mechanochemical coupling
Kata Sarlós et al. Proc Natl Acad Sci U S A. 2012.
Abstract
Maintenance of genome integrity is the major biological role of RecQ-family helicases via their participation in homologous recombination (HR)-mediated DNA repair processes. RecQ helicases exert their functions by using the free energy of ATP hydrolysis for mechanical movement along DNA tracks (translocation). In addition to the importance of translocation per se in recombination processes, knowledge of its mechanism is necessary for the understanding of more complex translocation-based activities, including nucleoprotein displacement, strand separation (unwinding), and branch migration. Here, we report the key properties of the ssDNA translocation mechanism of Escherichia coli RecQ helicase, the prototype of the RecQ family. We monitored the pre-steady-state kinetics of ATP hydrolysis by RecQ and the dissociation of the enzyme from ssDNA during single-round translocation. We also gained information on the translocation mechanism from the ssDNA length dependence of the steady-state ssDNA-activated ATPase activity. We show that RecQ occludes 18 ± 2 nt on ssDNA during translocation. The hydrolysis of ATP is noncooperative in the presence of ssDNA, indicating that RecQ active sites work independently during translocation. In the applied conditions, the enzyme hydrolyzes 35 ± 4 ATP molecules per second during ssDNA translocation. RecQ translocates at a moderate processivity, with a mean run length of 100-320 nt on ssDNA. The determined tight mechanochemical coupling of 1.1 ± 0.2 ATP consumed per nucleotide traveled indicates an inchworm-type mechanism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
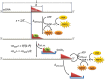
Fig. 1.
Model for RecQ translocation along ssDNA. (Top) RecQ (red triangle) binds to an ssDNA strand of track length L (expressed in nucleotides) and occludes a site of b nucleotides in length. (Middle) On ATP-driven translocation, the enzyme travels s nucleotides per ATP molecule consumed, defining the mechanochemical coupling ratio C (= 1/s; number of ATP molecules hydrolyzed per nucleotide traveled). RecQ performs ATPase activities of _k_ATP,trans and _k_ATP,end during translocation and on reaching the end of the DNA track, respectively. (Bottom) Enzyme dissociates at rate constants _k_off,int and _k_off,end from internal sites on DNA and the 5′-end, respectively. Processivity, expressed as the probability of performing the oncoming ATP-consuming translocation cycle as opposed to dissociation, is defined as P = _k_ATP,trans/(_k_ATP,trans + _k_off,int). The mean number of ATP molecules hydrolyzed (single-round ATP consumption amplitude) and nucleotides traveled (run length) in a processive run will thus be <_n_ATP> = P/(1 − P) and <_n_nt> = <_n_ATP>/C, respectively (15, 30, 31). In this study, we determined the above-mentioned parameters by using optical signals reporting the appearance of ATP hydrolysis products ADP (by NADH absorbance using a PK-LDH linked assay, orange) and Pi (by using a fluorescently labeled Pi binding protein, MDCC-PBP, yellow), as well as the intrinsic Trp fluorescence of the enzyme (green), which increases on dissociation from DNA. In single-round pre–steady-state experiments, we applied DxSO4 (blue) as a protein trap to prevent rebinding of RecQ to DNA after finishing the first processive run on ATP addition.
Fig. 2.
Single-round translocation of RecQ along ssDNA. (A) Time courses of Pi production from ATP during translocation of RecQ along ssDNA substrates of different length (dT12 − dT90, lengths in nucleotides indicated), which were recorded on mixing 25 nM RecQ plus 1 μM thymidine oligonucleotides of various length (dTn) with 0.5 mM ATP plus 0.04 mg/mL DxSO4 in the stopped flow. Pi production was monitored by using the fluorescence change of MDCC-PBP (5 μM in all syringes). (B) Oligo-dT length dependence of the corrected amplitudes of Pi production (mol Pi/mol RecQ) during the first phase, corresponding to translocation along ssDNA. Indicated values are DxSO4 concentrations in (μg/mL). Data were analyzed using
Eq. S1
. Data points were corrected based on
Fig. S2_B_
. (C) Dependence of the determined mean number of ATPase cycles performed in a processive run (<_n_ATP>; Fig. 1) on DxSO4 concentration during translocation along linear oligo-dT (black) and circular M13 phage ssDNA (gray). Data points obtained with dTn substrates were determined by extrapolation to infinite track length as shown in B. Data points obtained with dTn and M13 substrates at DxSO4 concentrations enabling effective trapping (
Fig. S2 A_–_E
) were used in fits based on
Eq. S2
(30) to determine the trap-free <_n_ATP> values. Obtained parameters are listed in Table 1.
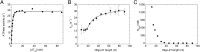
Fig. 3.
ssDNA length dependence of RecQ ATPase activity. (A) dT54 concentration dependence of the steady-state ATPase activity of RecQ. RecQ (15 nM) and ATP (1 mM) were titrated with increasing concentrations of dT54 in a PK-LDH coupled assay. Fit to the data using a quadratic binding equation yielded _K_DNA = 1.8 ± 0.1 nM and _k_cat = 29 ± 1 s−1 in the experiment shown. (B) Oligo-dT length dependence of _k_cat values determined from oligo-dT concentration dependence of the steady-state ATPase activity of RecQ. The solid line shows a fit based on
Eq. S3
(15). Obtained parameters are listed in Table 1. (C) Dependence of _K_DNA values (oligo-dT concentrations required for half saturation of ATPase activity of 15 nM RecQ) on oligo-dT length.
Fig. 4.
Dissociation of RecQ from ssDNA during single-round translocation. (A) Time courses of dissociation of 0.5 μM RecQ from 25 μM (concentration in nucleotides) poly-dT (pdT) on mixing with 5 mM ATP plus varying concentrations of DxSO4 (Lower to Upper: 0.05, 0.1, 0.2, and 0.5 mg/mL), monitored by the Trp fluorescence change of RecQ. Solid lines show single exponential fits to the data. (B) DxSO4 concentration dependence of the observed rate contants (_k_obs) of RecQ dissociation from pdT (experimental conditions were as in A; ■) or mixing RecQ plus pdT with 0.1 mg/mL DxSO4 in the absence of ATP (□). The intercept of the fitted linear indicated a trap-free dissociation rate constant (_k_off,int) of 0.12 ± 0.01 s−1 from internal ssDNA sites during translocation. AU, arbitrary units.
Similar articles
- Efficient coupling of ATP hydrolysis to translocation by RecQ helicase.
Rad B, Kowalczykowski SC. Rad B, et al. Proc Natl Acad Sci U S A. 2012 Jan 31;109(5):1443-8. doi: 10.1073/pnas.1119952109. Epub 2012 Jan 17. Proc Natl Acad Sci U S A. 2012. PMID: 22307597 Free PMC article. - Translocation of E. coli RecQ helicase on single-stranded DNA.
Rad B, Kowalczykowski SC. Rad B, et al. Biochemistry. 2012 Apr 3;51(13):2921-9. doi: 10.1021/bi3000676. Epub 2012 Mar 21. Biochemistry. 2012. PMID: 22409300 Free PMC article. - Processive translocation mechanism of the human Bloom's syndrome helicase along single-stranded DNA.
Gyimesi M, Sarlós K, Kovács M. Gyimesi M, et al. Nucleic Acids Res. 2010 Jul;38(13):4404-14. doi: 10.1093/nar/gkq145. Epub 2010 Mar 8. Nucleic Acids Res. 2010. PMID: 20211839 Free PMC article. - Insights into the Dynamics and Helicase Activity of RecD2 of Deinococcus radiodurans during DNA Repair: A Single-Molecule Perspective.
Islam F, Purkait D, Mishra PP. Islam F, et al. J Phys Chem B. 2023 May 25;127(20):4351-4363. doi: 10.1021/acs.jpcb.3c00778. Epub 2023 May 10. J Phys Chem B. 2023. PMID: 37163679 Review. - Ensemble methods for monitoring enzyme translocation along single stranded nucleic acids.
Tomko EJ, Fischer CJ, Lohman TM. Tomko EJ, et al. Methods. 2010 Jul;51(3):269-76. doi: 10.1016/j.ymeth.2010.03.010. Epub 2010 Apr 3. Methods. 2010. PMID: 20371288 Free PMC article. Review.
Cited by
- DNA helicases involved in DNA repair and their roles in cancer.
Brosh RM Jr. Brosh RM Jr. Nat Rev Cancer. 2013 Aug;13(8):542-58. doi: 10.1038/nrc3560. Epub 2013 Jul 11. Nat Rev Cancer. 2013. PMID: 23842644 Free PMC article. Review. - A nucleotide-dependent and HRDC domain-dependent structural transition in DNA-bound RecQ helicase.
Kocsis ZS, Sarlós K, Harami GM, Martina M, Kovács M. Kocsis ZS, et al. J Biol Chem. 2014 Feb 28;289(9):5938-49. doi: 10.1074/jbc.M113.530741. Epub 2014 Jan 8. J Biol Chem. 2014. PMID: 24403069 Free PMC article. - Shuttling along DNA and directed processing of D-loops by RecQ helicase support quality control of homologous recombination.
Harami GM, Seol Y, In J, Ferencziová V, Martina M, Gyimesi M, Sarlós K, Kovács ZJ, Nagy NT, Sun Y, Vellai T, Neuman KC, Kovács M. Harami GM, et al. Proc Natl Acad Sci U S A. 2017 Jan 24;114(4):E466-E475. doi: 10.1073/pnas.1615439114. Epub 2017 Jan 9. Proc Natl Acad Sci U S A. 2017. PMID: 28069956 Free PMC article. - The toposiomerase IIIalpha-RMI1-RMI2 complex orients human Bloom's syndrome helicase for efficient disruption of D-loops.
Harami GM, Pálinkás J, Seol Y, Kovács ZJ, Gyimesi M, Harami-Papp H, Neuman KC, Kovács M. Harami GM, et al. Nat Commun. 2022 Feb 3;13(1):654. doi: 10.1038/s41467-022-28208-9. Nat Commun. 2022. PMID: 35115525 Free PMC article. - Single molecule kinetics uncover roles for E. coli RecQ DNA helicase domains and interaction with SSB.
Bagchi D, Manosas M, Zhang W, Manthei KA, Hodeib S, Ducos B, Keck JL, Croquette V. Bagchi D, et al. Nucleic Acids Res. 2018 Sep 19;46(16):8500-8515. doi: 10.1093/nar/gky647. Nucleic Acids Res. 2018. PMID: 30053104 Free PMC article.
References
- Khakhar RR, Cobb JA, Bjergbaek L, Hickson ID, Gasser SM. RecQ helicases: Multiple roles in genome maintenance. Trends Cell Biol. 2003;13:493–501. - PubMed
- Bachrati CZ, Hickson ID. RecQ helicases: Guardian angels of the DNA replication fork. Chromosoma. 2008;117:219–233. - PubMed
- Wu L, Hickson ID. DNA helicases required for homologous recombination and repair of damaged replication forks. Annu Rev Genet. 2006;40:279–306. - PubMed
- Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials