Differential anti-proliferative activities of poly(ADP-ribose) polymerase (PARP) inhibitors in triple-negative breast cancer cells - PubMed (original) (raw)
Differential anti-proliferative activities of poly(ADP-ribose) polymerase (PARP) inhibitors in triple-negative breast cancer cells
Hsiao-Ching Chuang et al. Breast Cancer Res Treat. 2012 Jul.
Abstract
Despite recent advances in the clinical evaluation of various poly(ADP-ribose) polymerase (PARP) inhibitors in triple-negative breast cancer (TNBC) patients, data defining potential anti-tumor mechanisms beyond PARP inhibition for these agents are lacking. To address this issue, we investigated the effects of four different PARP inhibitors (AG-014699, AZD-2281, ABT-888, and BSI-201) in three genetically distinct TNBC cell lines (MDA-MB-468, MDA-MB-231, and Cal-51). Assays of cell viability and colony formation and flow cytometric analysis were used to determine effects on cell growth and cell cycle progression. PARP-dependent and -independent signaling mechanisms of each PARP inhibitor were investigated by western blotting and shRNA approaches. Potential synergistic interactions between PARP inhibitors and cisplatin in suppressing TNBC cell viability were assessed. These PARP inhibitors exhibited differential anti-tumor activities, with the relative potencies of AG-014699 > AZD-2281 > ABT-888 > BSI-201. The higher potencies of AG-014699 and AZD-2281 were associated with their effects on G(2)/M arrest and DNA damage as manifested by γ-H2AX formation and, for AG-014699, its unique ability to suppress Stat3 phosphorylation. Abilities of individual PARP inhibitors to sensitize TNBC cells to cisplatin varied to a great extent in a cell context- and cell line-specific manner. Differential activation of signaling pathways suggests that the PARP inhibitors currently in clinical trials have different anti-tumor mechanisms beyond PARP inhibition and these PARP-independent mechanisms warrant further investigation.
Conflict of interest statement
Conflict of interest The authors declare no conflict of interest.
Figures
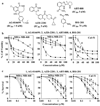
Fig. 1
Differential effects of PARP inhibitors on the viability and survival of TNBC cells. a Structures and reported IC50 values for PARP inhibition of AG-014699 [15], AZD-2281 [16], ABT-888 [17], and BSI-201 [18]. b MTT assays of the dose-dependent suppressive effects of AG-014699, AZD-2281, ABT-888, and BSI-201 on the viability of MDA-MB-468, MDA-MB-231, and Cal-51 cells in 5 % FBS-supplemented DMEM medium after 72 h of treatment. Mean ± SD (n = 6). c Clonogenic assays of the dose-dependent suppressive effects of AG-014699, AZD-2281, ABT-888, and BSI-201 on the survival of MDA-MB-468, MDA-MB-231, and Cal-51 cells in 5 % FBS-supplemented DMEM medium after 14–21 days of treatment. Mean ± SD (n = 3)
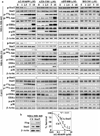
Fig. 2
Differential effects of PARP inhibitors on PARP inhibition, DNA damage, and apoptosis in TNBC cells. Effects of AG-014699, AZD-2281, ABT-888, or BSI-201 on the levels of PAR, γ-H2AX, and PARP cleavage in MDA-MB-468, MDA-MB-231, and Cal-51 cells. Cells were treated with the indicated concentrations of PARP inhibitors in the presence of 5 % FBS-supplemented DMEM medium for 72 h
Fig. 3
Differential effects of PARP inhibitors on the activation status of signaling effectors pertinent to cell proliferation and survival in TNBC cells, suggesting the heterogeneity in their modes of action, a Effects of AG-014699, AZD-2281, ABT-888, or BSI-201 on the phosphorylation status of Stat3, Akt, ERK, and p38 in MDA-MB-468, MDA-MB-231, and Cal-51 cells after 72 h of treatment, bLeft, verification by western blotting of ectopic expression of Stat3 in MDA-MB-468 cells transfected with an empty pcDNA vector or plasmid encoding Flag-tagged constitutively active Stat3 (CA-Stat3). Right, effect of the ectopic expression of CA-Stat3 versus the pcDNA vector on AG-014699-mediated suppression of clonogenic survival of MDA-MB-468 cells. Mean ± SD (n = 6). * P < 0.05
Fig. 4
AG-014699- and AZD-2281-mediated activation of Akt and ERK might be attributable to downregulation of PHLPP expression in MDA-MB-468 cells, a Dose-dependent effects of AG-014699 and AZD-2281 versus ABT-888 on PHLPP expression after 72 h of treatment, b Effect of LY-294002 (10 µM) on AG-014699- and AZD-2281-mediated phosphorylation of Ser473-Akt and ERKs
Fig. 5
Effect of shRNA-mediated silencing of p53 on the sensitivity of Cal-51 cells to the anti-proliferative effects of PARP inhibitors, a Verification by western blotting of shRNA-mediated knockdown of p53 expression in the stably transfected Cal-51 subclone. b Effects of p53 knockdown on the potencies of AG-014699, AZD-2281, ABT-888, and BSI-201 in suppressing the viability of Cal-51 cells after 72 h of treatment. Mean ± SD (n = 6). * P< 0.05. c Effects of p53 knockdown on the potencies of AG-014699, AZD-2281, ABT-888, and BSI-201 in suppressing the clonogenic survival of Cal-51 cells. Mean ± SD (n = 3). * P<0.05. d Effects of p53 knockdown on the DNA damage response, as indicated by γ-H2AX formation, to AG-014699 and AZD-2281
Fig. 6
Sensitization of TNBC cells to cisplatin-mediated cell death and DNA damage by PARP inhibitors, a Differential susceptibility of MDA-MB-468 (468), MDA-MB-231 (231), and Cal-51 (51) cells to cisplatin-mediated suppression of cell viability after 72 h of treatment. Mean ± SD (n = 6). b Effects of AZD-2281, AG-014699, ABT-888, and BSI-201 on the sensitivity of MDA-MB-468 cells to cisplatin. Mean ± SD (n = 6). § CI > 1; ≠ CI = 1; * 1 > CI > 0.5; ** CI < 0.5. c Western blot analysis of the effects of AZD-2281, AG-014699, ABT-888, and BSI-201 on the sensitivity of MDA-MB-468 cells to cisplatin-induced DNA damage, as indicated by γ-H2AX formation, d Effects of AZD-2281, AG-014699, ABT-888, and BSI-201 on the sensitivity of Cal-51 and MDA-MB-231 cells to cisplatin. Mean ± SD (n = 6). e Western blot analysis of the effects of AZD-2281, AG-014699, ABT-888, and BSI-201 on the sensitivity of Cal-51 and MDA-MB-231 cells to cisplatin-induced DNA damage, as indicated by γ-H2AX formation
Similar articles
- Co-targeting poly(ADP-ribose) polymerase (PARP) and histone deacetylase (HDAC) in triple-negative breast cancer: Higher synergism in BRCA mutated cells.
Marijon H, Lee DH, Ding L, Sun H, Gery S, de Gramont A, Koeffler HP. Marijon H, et al. Biomed Pharmacother. 2018 Mar;99:543-551. doi: 10.1016/j.biopha.2018.01.045. Epub 2018 Feb 20. Biomed Pharmacother. 2018. PMID: 29902865 - Comparative antiproliferative effects of iniparib and olaparib on a panel of triple-negative and non-triple-negative breast cancer cell lines.
Pierce A, McGowan PM, Cotter M, Mullooly M, O'Donovan N, Rani S, O'Driscoll L, Crown J, Duffy MJ. Pierce A, et al. Cancer Biol Ther. 2013 Jun;14(6):537-45. doi: 10.4161/cbt.24349. Cancer Biol Ther. 2013. PMID: 23760496 Free PMC article. - Response of subtype-specific human breast cancer-derived cells to poly(ADP-ribose) polymerase and checkpoint kinase 1 inhibition.
Shibata H, Miuma S, Saldivar JC, Huebner K. Shibata H, et al. Cancer Sci. 2011 Oct;102(10):1882-8. doi: 10.1111/j.1349-7006.2011.02016.x. Epub 2011 Jul 21. Cancer Sci. 2011. PMID: 21707865 Free PMC article. - Triple-negative breast cancer and poly(ADP-ribose) polymerase inhibitors.
Park Y, Moriyama A, Kitahara T, Yoshida Y, Urita T, Kato R. Park Y, et al. Anticancer Agents Med Chem. 2012 Jul;12(6):672-7. doi: 10.2174/187152012800617759. Anticancer Agents Med Chem. 2012. PMID: 22263793 Review. - The antimitotic potential of PARP inhibitors, an unexplored therapeutic alternative.
Iglesias P, Costoya JA. Iglesias P, et al. Curr Top Med Chem. 2014;14(20):2346-65. doi: 10.2174/1568026614666141130100641. Curr Top Med Chem. 2014. PMID: 25434352 Review.
Cited by
- PARP inhibitors: current status and implications for anticancer therapeutics.
Usmani H, Hussain SA, Sheikh A. Usmani H, et al. Infect Agent Cancer. 2013 Dec 2;8(1):46. doi: 10.1186/1750-9378-8-46. Infect Agent Cancer. 2013. PMID: 24289880 Free PMC article. - INPP4B overexpression enhances the antitumor efficacy of PARP inhibitor AG014699 in MDA-MB-231 triple-negative breast cancer cells.
Sun Y, Ding H, Liu X, Li X, Li L. Sun Y, et al. Tumour Biol. 2014 May;35(5):4469-77. doi: 10.1007/s13277-013-1589-y. Epub 2014 Jan 14. Tumour Biol. 2014. PMID: 24420152 - Novel drug targets for personalized precision medicine in relapsed/refractory diffuse large B-cell lymphoma: a comprehensive review.
Camicia R, Winkler HC, Hassa PO. Camicia R, et al. Mol Cancer. 2015 Dec 11;14:207. doi: 10.1186/s12943-015-0474-2. Mol Cancer. 2015. PMID: 26654227 Free PMC article. Review. - Small-Molecule Inhibitors Targeting DNA Repair and DNA Repair Deficiency in Research and Cancer Therapy.
Hengel SR, Spies MA, Spies M. Hengel SR, et al. Cell Chem Biol. 2017 Sep 21;24(9):1101-1119. doi: 10.1016/j.chembiol.2017.08.027. Cell Chem Biol. 2017. PMID: 28938088 Free PMC article. Review. - Synergistic Effect of Trabectedin and Olaparib Combination Regimen in Breast Cancer Cell Lines.
Ávila-Arroyo S, Nuñez GS, García-Fernández LF, Galmarini CM. Ávila-Arroyo S, et al. J Breast Cancer. 2015 Dec;18(4):329-38. doi: 10.4048/jbc.2015.18.4.329. Epub 2015 Dec 23. J Breast Cancer. 2015. PMID: 26770239 Free PMC article.
References
- Reis-Filho JS, Tutt AN. Triple negative tumours: a critical review. Histopathology. 2008;52(1):108–118. - PubMed
- Carey L, Winer E, Viale G, Cameron D, Gianni L. Triple-negative breast cancer: disease entity or title of convenience? Nat Rev Clin Oncol. 2010;7(12):683–692. - PubMed
- Carey LA, Dees EC, Sawyer L, Gatti L, Moore DT, Collichio F, Ollila DW, Sartor CI, Graham ML, Perou CM. The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res. 2007;13(8):2329–2334. - PubMed
- Gonzalez-Angulo AM, Timms KM, Liu S, Chen H, Litton JK, Potter J, Lanchbury JS, Stemke-Hale K, Hennessy BT, Aran BK, Hortobagyi GN, Do KA, Mills GB, Meric-Bernstam F. Incidence and outcome of BRCA mutations in unselected patients with triple receptor-negative breast cancer. Clin Cancer Res. 2011;17(5):1082–1089. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous