Human gut microbiome viewed across age and geography - PubMed (original) (raw)
Comparative Study
. 2012 May 9;486(7402):222-7.
doi: 10.1038/nature11053.
Federico E Rey, Mark J Manary, Indi Trehan, Maria Gloria Dominguez-Bello, Monica Contreras, Magda Magris, Glida Hidalgo, Robert N Baldassano, Andrey P Anokhin, Andrew C Heath, Barbara Warner, Jens Reeder, Justin Kuczynski, J Gregory Caporaso, Catherine A Lozupone, Christian Lauber, Jose Carlos Clemente, Dan Knights, Rob Knight, Jeffrey I Gordon
Affiliations
- PMID: 22699611
- PMCID: PMC3376388
- DOI: 10.1038/nature11053
Comparative Study
Human gut microbiome viewed across age and geography
Tanya Yatsunenko et al. Nature. 2012.
Abstract
Gut microbial communities represent one source of human genetic and metabolic diversity. To examine how gut microbiomes differ among human populations, here we characterize bacterial species in fecal samples from 531 individuals, plus the gene content of 110 of them. The cohort encompassed healthy children and adults from the Amazonas of Venezuela, rural Malawi and US metropolitan areas and included mono- and dizygotic twins. Shared features of the functional maturation of the gut microbiome were identified during the first three years of life in all three populations, including age-associated changes in the genes involved in vitamin biosynthesis and metabolism. Pronounced differences in bacterial assemblages and functional gene repertoires were noted between US residents and those in the other two countries. These distinctive features are evident in early infancy as well as adulthood. Our findings underscore the need to consider the microbiome when evaluating human development, nutritional needs, physiological variations and the impact of westernization.
Figures
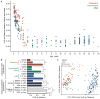
Fig. 1. Differences in the fecal microbial communities of Malawians, Amerindians and residents of the USA at different ages
(a) UniFrac distances between children and adults decrease with increasing age of children in each population. Each point shows an average distance between a child and all adults unrelated to that child but from the same country. Results are derived from bacterial V4-16S rRNA datasets. (b) Large interpersonal variations are observed in the phylogenetic configurations of fecal microbial communities at early ages. Malawian and Amerindian children and adults are more similar to one another than to USA children and adults. UniFrac distances were defined from bacterial V4-16S rRNA data generated from the microbiota of 181 unrelated adults (≥18 years old) and 204 unrelated children (n=31 Malawians 0.03–3 years old, 21 3–17 years old; 30 Amerindians 0.08–3 years old, 29 3–17 years old; 31 residents of the USA 0.08–3 years old, 62 sampled at 3–17 years of age). Abbreviations: * p<0.05; **p<0.005 (Student’s t-test with 1000 Monte Carlo simulations). See Table S3 for a complete description of the statistical significance of all comparisons shown in the Figure. (c) PCoA of unweighted UniFrac distances for the fecal microbiota of adults.
Fig. 2. Bacterial diversity increases with age in each population
Number of observed 97%ID OTUs plotted against age for (a) all subjects, (b) during the first 3 years of life, and (c) adults (in the latter panel, average values ± SEM are plotted). *p<0.05, **p<0.005 (ANOVA with Bonferroni post-hoc test).
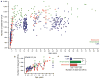
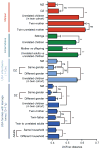
Fig. 3. Differences in the functional profiles of fecal microbiomes in the three study populations
Examples of KEGG ECs that exhibited the largest differences, as determined by Random Forests and ShotgunFuntionalizeR analyses, in proportional representation between USA and Malawian/Amerindian populations. Shown are the relative abundances of genes encoding the indicated ECs (normalized by Z-score across all datasets). (a) UPGMA clustering of 10 USA, 10 Malawian and 6 Amerindian baby fecal microbiomes. (b) UPGMA clustering of 16 USA, 5 Malawian and 5 Amerindian adult fecal microbiomes.
Fig. 4. Differences in the fecal microbiota between family members across the three populations studied
UniFrac distances between the fecal bacterial communities of family members were calculated (n=19 Amerindian, 34 Malawian and 54 USA families with teenage twins). Mean ± SEM values are plotted. The UniFrac matrix was permutated 1000 times; p values represent the fraction of times permuted differences were greater than real differences: ns (not significant; p>0.05), * p<0.05, **p<0.005.
References
Publication types
MeSH terms
Substances
Grants and funding
- K05 AA017688/AA/NIAAA NIH HHS/United States
- P01 DK078669/DK/NIDDK NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- T32-HD049338/HD/NICHD NIH HHS/United States
- DK078669/DK/NIDDK NIH HHS/United States
- T32 HD049338/HD/NICHD NIH HHS/United States
- K01 DK090285/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases