A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3 - PubMed (original) (raw)
A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3
Pablo Cingolani et al. Fly (Austin). 2012 Apr-Jun.
Abstract
We describe a new computer program, SnpEff, for rapidly categorizing the effects of variants in genome sequences. Once a genome is sequenced, SnpEff annotates variants based on their genomic locations and predicts coding effects. Annotated genomic locations include intronic, untranslated region, upstream, downstream, splice site, or intergenic regions. Coding effects such as synonymous or non-synonymous amino acid replacement, start codon gains or losses, stop codon gains or losses, or frame shifts can be predicted. Here the use of SnpEff is illustrated by annotating ~356,660 candidate SNPs in ~117 Mb unique sequences, representing a substitution rate of ~1/305 nucleotides, between the Drosophila melanogaster w(1118); iso-2; iso-3 strain and the reference y(1); cn(1) bw(1) sp(1) strain. We show that ~15,842 SNPs are synonymous and ~4,467 SNPs are non-synonymous (N/S ~0.28). The remaining SNPs are in other categories, such as stop codon gains (38 SNPs), stop codon losses (8 SNPs), and start codon gains (297 SNPs) in the 5'UTR. We found, as expected, that the SNP frequency is proportional to the recombination frequency (i.e., highest in the middle of chromosome arms). We also found that start-gain or stop-lost SNPs in Drosophila melanogaster often result in additions of N-terminal or C-terminal amino acids that are conserved in other Drosophila species. It appears that the 5' and 3' UTRs are reservoirs for genetic variations that changes the termini of proteins during evolution of the Drosophila genus. As genome sequencing is becoming inexpensive and routine, SnpEff enables rapid analyses of whole-genome sequencing data to be performed by an individual laboratory.
Figures
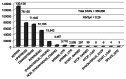
Figure 1. Classification of SNPs in _w_1118; _iso-_2; _iso_-3. The number of NSPs in each class is shown above the bar. The quality score was arbitrarily set at 70 and above for this graph.
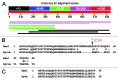
Figure 2. Analysis of Eip63E start-gained SNP in _w_1118; _iso-_2; _iso_-3. (A) Location of the start-gained SNP at the Eip63E locus. Notice that the reading frame is the same as the normal translation start site (TSS). (B) Conservation of 60 amino acid N-terminal region of Eip63E in _w_1118; _iso-_2; _iso_-3 with Drosophila yakuba orthologous gene. The other sequenced Drosophila species do not have this N-terminal sequence (not shown).
Figure 3. Oc/Otd has two stop-gained SNPs in _w_1118; _iso-_2; _iso_-3. (A) Location of the two stop gained SNPs in oc/otd. (B) Protein BLAST of Oc/Otd against the non-redundant (nr) protein database shows that only the 60 amino Hox domain flanking amino acid 100 is conserved from Drosophila to humans. The color coding shows the alignment scores.
Figure 4. CG34326 has one stop-gained SNP in _w_1118; _iso-_2; _iso_-3 in the non-conserved C-terminal region. (A) Protein BLAST of CG34326 against the non-redundant (nr) protein database shows that only the 38 N-terminal amino acids are conserved among Drosophila species and not beyond Drosophila. The colored lines represent the homologs from the following organisms: Drosophila melanogaster, Drosophila grimshawi, Drosophila yakuba, Drosophila erecta, Drosophila virilus, Ixodes scapularis, Ixodes scapularis, and Nycticebus coucang. (B) Aligment of Drosophila melanogaster CG34326 with orthologous gene from Drosophila grimshawi. (C) Aligment of Drosophila melanogaster CG34326 with orthologous gene from Drosophila yakuba.
Figure 5. CG13958 has a stop lost SNP in _w_1118; _iso-_2; _iso_-3. The top comparison shows the alignment of the Drosophila melanogaster reference genome with _w_1118; _iso-_2; _iso_-3. Notice that the stop lost causes an extension of nine amino acids. The second through sixth comparisons shows the alignment of Drosophila simulans, Drosophila erecta, Drosophila yakuba, Drosophila mojavensis, and Drosophila pseudoobscura pseudoobscura (Sbjct) with the Drosophila melanogaster reference genome (Dm-ref). The number of terminal amino acids missing or gained is shown (-1 to +3).
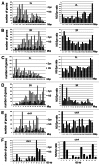
Figure 6. Nonsynonymous to synonymous ratios along the chromosome arms in _w_1118; _iso-_2; _iso_-3. (A) Left, Nonsynonymous SNPs at 1 Mbp intervals along the 2L chromosome arm (black) and synonymous SNPs (gray). Right, N/S ratios (NS/Syn) along the chromosome arms. Notice that N/S ratios are higher near the centromere and telomere (see text). (B-F) as in (A), but for chromosome arms 2R, 3L, 3R, 4 and X.
Similar articles
- Massively parallel resequencing of the isogenic Drosophila melanogaster strain w(1118); iso-2; iso-3 identifies hotspots for mutations in sensory perception genes.
Platts AE, Land SJ, Chen L, Page GP, Rasouli P, Wang L, Lu X, Ruden DM. Platts AE, et al. Fly (Austin). 2009 Jul-Sep;3(3):192-203. doi: 10.4161/fly.3.3.9652. Epub 2009 Jul 23. Fly (Austin). 2009. PMID: 19690466 Free PMC article. - Genome features of "Dark-fly", a Drosophila line reared long-term in a dark environment.
Izutsu M, Zhou J, Sugiyama Y, Nishimura O, Aizu T, Toyoda A, Fujiyama A, Agata K, Fuse N. Izutsu M, et al. PLoS One. 2012;7(3):e33288. doi: 10.1371/journal.pone.0033288. Epub 2012 Mar 14. PLoS One. 2012. PMID: 22432011 Free PMC article. - Drosophila melanogaster: a case study of a model genomic sequence and its consequences.
Ashburner M, Bergman CM. Ashburner M, et al. Genome Res. 2005 Dec;15(12):1661-7. doi: 10.1101/gr.3726705. Genome Res. 2005. PMID: 16339363 Review. - Evolutionary insights from large scale resequencing datasets in Drosophila melanogaster.
Guirao-Rico S, González J. Guirao-Rico S, et al. Curr Opin Insect Sci. 2019 Feb;31:70-76. doi: 10.1016/j.cois.2018.11.002. Epub 2018 Nov 20. Curr Opin Insect Sci. 2019. PMID: 31109676 Review.
Cited by
- Genetic marker: a genome mapping tool to decode genetic diversity of livestock animals.
Panchariya DC, Dutta P, Ananya, Mishra A, Chawade A, Nayee N, Azam S, Gandham RK, Majumdar S, Kushwaha SK. Panchariya DC, et al. Front Genet. 2024 Oct 17;15:1463474. doi: 10.3389/fgene.2024.1463474. eCollection 2024. Front Genet. 2024. PMID: 39483851 Free PMC article. Review. - Biallelic GGGCC repeat expansion leading to NAXE-related mitochondrial encephalopathy.
Ozaki K, Yatsuka Y, Oyazato Y, Nishiyama A, Nitta KR, Kishita Y, Fushimi T, Shimura M, Noma S, Sugiyama Y, Tagami M, Fukunaga M, Kinoshita H, Hirata T, Suda W, Murakawa Y, Carninci P, Ohtake A, Murayama K, Okazaki Y. Ozaki K, et al. NPJ Genom Med. 2024 Oct 25;9(1):48. doi: 10.1038/s41525-024-00429-5. NPJ Genom Med. 2024. PMID: 39455596 Free PMC article. - Outcomes of bone marrow mononuclear cell transplantation combined with interventional education for autism spectrum disorder.
Nguyen Thanh L, Nguyen HP, Ngo MD, Bui VA, Dam PTM, Bui HTP, Ngo DV, Tran KT, Dang TTT, Duong BD, Nguyen PAT, Forsyth N, Heke M. Nguyen Thanh L, et al. Stem Cells Transl Med. 2021 Jan;10(1):14-26. doi: 10.1002/sctm.20-0102. Epub 2020 Sep 9. Stem Cells Transl Med. 2021. PMID: 32902182 Free PMC article. Clinical Trial. - Genetic variation at the Cyp6m2 putative insecticide resistance locus in Anopheles gambiae and Anopheles coluzzii.
Wagah MG, Korlević P, Clarkson C, Miles A; Anopheles gambiae 1000 Genomes Consortium; Lawniczak MKN, Makunin A. Wagah MG, et al. Malar J. 2021 May 25;20(1):234. doi: 10.1186/s12936-021-03757-4. Malar J. 2021. PMID: 34034756 Free PMC article. - Genomic signals found using RNA sequencing show signatures of selection and subtle population differentiation in walleye (Sander vitreus) in a large freshwater ecosystem.
Thorstensen MJ, Jeffrey JD, Treberg JR, Watkinson DA, Enders EC, Jeffries KM. Thorstensen MJ, et al. Ecol Evol. 2020 Jun 13;10(14):7173-7188. doi: 10.1002/ece3.6418. eCollection 2020 Jul. Ecol Evol. 2020. PMID: 32760520 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
- P30 ES06639/ES/NIEHS NIH HHS/United States
- R01 ES012933/ES/NIEHS NIH HHS/United States
- P30 ES006639/ES/NIEHS NIH HHS/United States
- R01 DK071073/DK/NIDDK NIH HHS/United States
- R21 ES021983/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous